A Quick Overview of Molecular Cloning
by Tyasning Kroemer, Ph.D.

by Tyasning Kroemer, Ph.D.
Molecular cloning allows us to study genes, modify genes, reintroduce modified genes into the natural host or into another host, or to overexpress a recombinant protein.
You can perform molecular cloning by incorporating a piece of DNA into a vector, such as a plasmid (a recombinant DNA) to make more identical recombinant DNA in a living host.
Things to consider before you clone are the availability of your cloning materials, the goal of your research, and the difficulty level of your protocol.
To begin with, you have to prepare some important materials for cloning:
The source for a DNA fragment or “DNA insert” can be genomic DNA, complementary DNA, plasmid DNA, PCR product, or synthetic DNA. The DNA insert must contain particular sequences at the end of the fragments compatible with the prepared vector. You can add these particular sequences onto your DNA insert by PCR.
A vector is a DNA molecule which can carry a DNA insert to generate a recombinant DNA and replicate in a particular host.
Some examples of vectors are:

Elements in a plasmid vector:
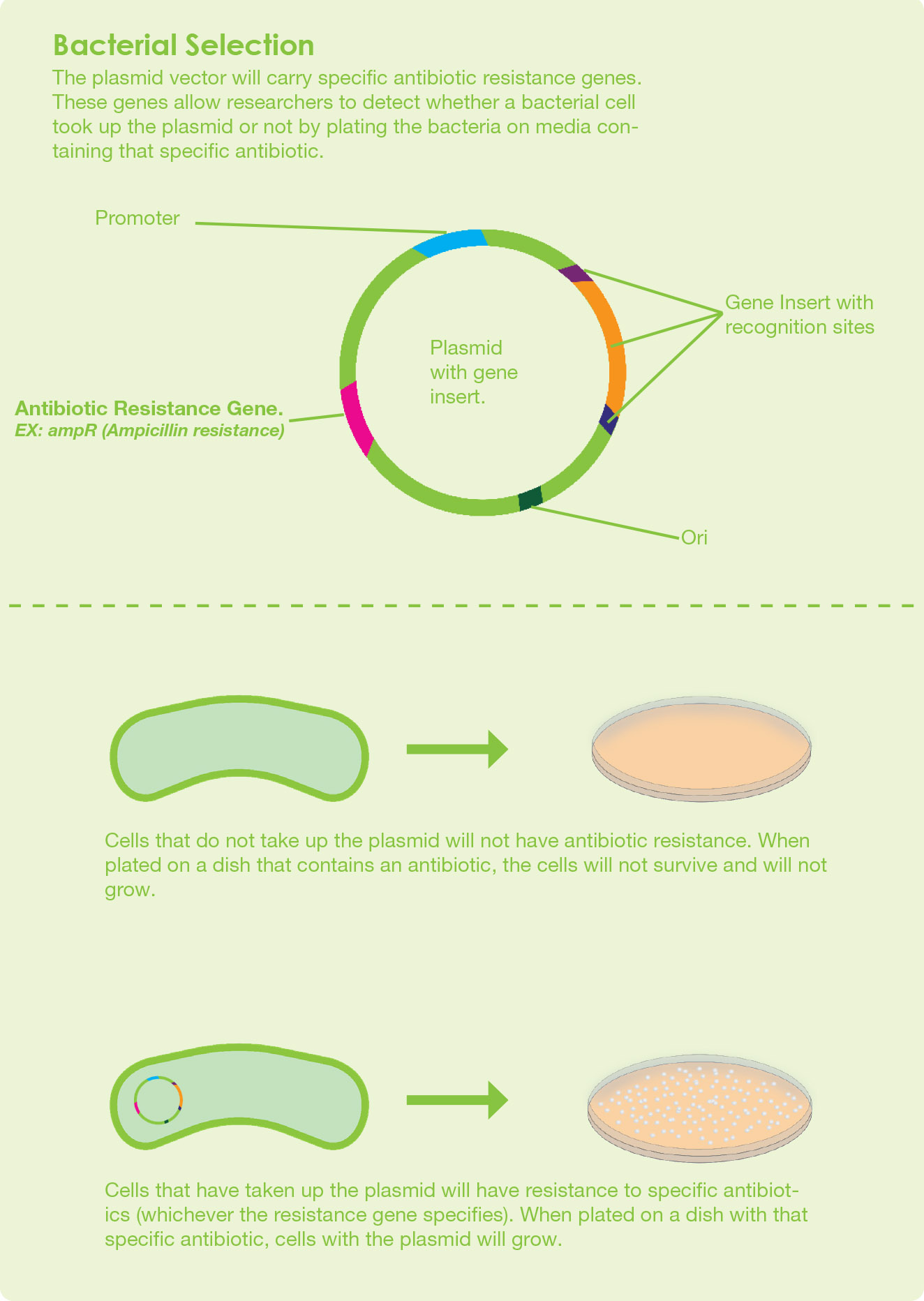
After a DNA fragment is incorporated into the plasmid vector, the next cloning step is to perform a transformation step. In this transformation step, the recombinant DNA is introduced into the competent cell by a chemical process or by electroporation. Competent cells are cells that are temporarily permeable to extracellular DNA. The host organisms which are commonly used in the laboratories are Escherichia coli and Saccharomyces cerevisiae
Selective medium is a growth medium containing a selective agent to grow the transformed host.
When you choose antibiotic selection for cloning, your growth medium must contain antibiotics. The antibiotics should correspond to the antibiotic selectable marker gene on the vector.
The most common antibiotics used for selection are Ampicillin, Kanamycin, and Chloramphenicol.
There are several different approaches to clone and you will need to find the right approach for your research. Below are some examples of popular cloning methods to generate a recombinant DNA construct:
Restriction enzymes are enzymes which cut DNA near at a specific short nucleotide sequence called a restriction site. The restriction enzyme based cloning method depends on the activity of restriction enzymes to ‘cut’ both a vector and a DNA insert and the method also depends on a DNA ligase to ‘paste’ the DNA fragment into the vector. This method is useful when have one DNA insert to incorporate into the plasmid.
By using PCR, you can add restriction enzyme sites on your DNA insert to accommodate this method. Your DNA insert must not contain an internal restriction site similar to the restriction site on your plasmid. Your restriction enzyme can cut your DNA insert at this internal restriction site and produce unwanted smaller pieces of DNA fragments.
You can choose to use one restriction enzyme or two enzymes to cut your DNA fragment and vector. When using two enzymes, both enzymes must be compatible or work well in the same restriction enzyme buffer.

Restriction Enzyme Based Cloning. 1. Short sequences containing restriction sites are added into the 5’ ends of primers for DNA amplification by PCR. 2. Both the vector and DNA fragment are digested with restriction enzymes to create cohesive ends. 3. The vector and DNA fragment are ligated. 4. The recombinant DNA enters the host cell during transformation.
PCR cloning relies on a process called ligation, which is a method of inserting a DNA fragment into a vector using DNA ligase. The reason ligation is important for this step is because it is responsible for inserting the PCR product into a ‘T-tailed’ plasmid.
PCR amplified inserts contain an adenine residue at the 3’ end of the DNA fragments (‘A-tailed’ ends). A ‘T-tailed’ plasmid vector has a single 3’ deoxythymidine (T) at each end of the arms of a linearized plasmid. Therefore, these PCR products can be ligated into ‘T-tailed’ vectors by using DNA ligase, and this step is followed by transformation.
You can choose this method when your restriction enzymes are not compatible or you find an internal restriction enzyme site in your DNA insert.
One disadvantage of this method is you will need a specific T-tailed vector to perform PCR cloning. But T-tailed vectors may not have supportive elements such as promoter region or protein tag which are needed to express your intended recombinant protein.

PCR Cloning. 1. PCR Product with A-tailed ends is combined with T-tailed vector. 2. During ligation, PCR product is inserted into the vector.
Ligation independent cloning (LIC) is performed by generating short sequences at the end of a DNA insert that matches the short sequences of a plasmid vector. Enzymes with 3’ to 5’ exonuclease activity chew 3’ ends and generate cohesive ends between the DNA fragment and the linearized vector. The two materials are then combined for annealing step. During transformation, the host organism repairs the nicks on the recombinant DNA.
The advantage of this method is it won’t create any new restriction sites or unwanted sequences in the final DNA construct.

LIC Cloning. 1. Short sequences which matches with sequences on the plasmid are added into the 5’ ends of primers for DNA amplification by PCR. 2. Plasmid is linearized by using restriction enzyme. 3. Both DNA insert and vector are treated with 3’ to 5’ exonuclease to create cohesive overhangs. 4. Both DNA and vector are annealed. 5. After transformation, the host cell repairs the nicks on the recombinant DNA.
The seamless cloning (SC) technique (similar to LIC) depends on matching short sequences at the ends of a DNA fragment to the short sequences on a plasmid vector. SC method requires an enzyme with 5’ to 3’ exonuclease activity to create 3’ overhangs, a DNA polymerase to fill in gaps, and a DNA ligase to seal the nicks.
The advantage of LIC and SC over the restriction enzyme-based cloning is it allows insertion of more than one DNA fragment into a vector. In addition, when you find an internal restriction enzyme site on your DNA fragment, you can use LIC or SC as an optional cloning method.

Seamless Cloning. 1. Short sequences are added into the 5’ ends of primers for DNA amplification by PCR. 2. Vector is digested by a restriction enzyme. 3. Both DNA fragment and vector are treated with an enzyme with 5’ to 3’ exonuclease activity to create cohesive overhangs. 4. During ligation, the DNA fragment is inserted into the vector.
This method requires site-specific DNA recombinase enzymes, which exchange and recombine DNA pieces with particular recombination sites.
The first step in this method is to insert a DNA fragment into an entry vector generating an entry clone. Another way to create an entry clone is by swapping and recombining a donor vector into an entry clone.
After creating an entry clone, the next step is to swap and recombine the entry clone into a destination clone. The benefit of this approach is it can be used to place more than five elements into a single vector. It is commonly used to identify protein-binding interactions or to optimize protein expression, purification and solubility. To perform this method, you will need a particular plasmid which has recombination sites.

Recombinational Cloning. 1. DNA fragment is inserted into an entry vector to create an entry clone. 2. Entry clone and destination vector are combined by a recombinase enzyme to create a destination clone.
In this article, we briefly explained five molecular cloning methods which are commonly used by researchers. Molecular cloning has been used for many different purposes in a variety of research fields. It is a fundamental steppingstone in modern biotechnology, whereby a new DNA fragment is first inserted in a vector, then the vector with the insert is inserted into a host cell. The entire process has been scaled up to the whole organismal level in mice and plants, for example.
In agriculture, cloning can shorten the time required to insert and develop a new beneficial trait in crops, such as a drought tolerant trait or a pest resistant trait. On the other hand, molecular cloning can be used to produce therapeutic proteins, such as a clot dissolving protein and interferon, or to synthesize other useful proteins, such as insulin, growth hormones, and monoclonal antibodies.
Overall, the application of molecular cloning continues to improve and creates more remarkable developments in the fields of agriculture, pharmaceutical industry, and biomedical research.
Amarakoon, I. I., Hamilton, C. L., Mitchell, S. A., Tennant, P. F., & Roye, M. E. (2017). Chapter 28 - Biotechnology. In S. Badal & R. Delgoda (Eds.), Pharmacognosy (pp. 549-563). Boston: Academic Press.
Bertero, A., Brown, S., & Vallier, L. (2017). Chapter 2 - Methods of Cloning. In M. Jalali, F. Y. L. Saldanha, & M. Jalali (Eds.), Basic Science Methods for Clinical Researchers (pp. 19-39). Boston: Academic Press.
Carter, M., & Shieh, J. C. (2010). Chapter 9 - Molecular Cloning and Recombinant DNA Technology. In M. Carter & J. C. Shieh (Eds.), Guide to Research Techniques in Neuroscience (pp. 207-227). New York: Academic Press.
Celie, P. H. N., Parret, A. H. A., & Perrakis, A. (2016). Recombinant cloning strategies for protein expression. Current Opinion in Structural Biology, 38, 145-154. doi: 10.1016/j.sbi.2016.06.010.
Gibson, D. G., Young, L., Chuang, R.-Y., Venter, J. C., Hutchison, C. A., & Smith, H. O. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6(5), 343-345. doi:10.1038/nmeth.1318.
Griffiths A.J.F., Miller J.H., & Suzuki D.T., e. a. (2000). Cloning a specific gene. New York: W. H. Freeman and Company.
Lessard, J. C. (2013). Molecular cloning. Methods Enzymol, 529, 85-98. doi:10.1016/b978-0-12-418687-3.00007-0
Li, M. Z., & Elledge, S. J. (2012). SLIC: A Method for Sequence- and Ligation-Independent Cloning. In J. Peccoud (Ed.), Gene Synthesis: Methods and Protocols (pp. 51-59). Totowa, NJ: Humana Press.
Lodish H., Berk A., Zipursky S.L., & al., e. (2000). DNA Cloning with Plasmid Vectors. New York: W. H. Freeman.
Park, J., Throop, A. L., & LaBaer, J. (2015). Site-Specific Recombinational Cloning Using Gateway and In-Fusion Cloning Schemes. Current protocols in molecular biology, 110(1), 3.20.21-23.20.23. doi:10.1002/0471142727.mb0320s110.
National Research Council Committee (1987). In Agricultural Biotechnology: Strategies for National Competitiveness. Washington (DC): National Academies Press (US) Copyright (c) National Academy of Sciences.
Trower, M. K., & Elgar, G. S. (1994). PCR cloning using T-vectors. Methods Mol Biol, 31, 19-33. doi:10.1385/0-89603-258-2:19.

Covalently conjugating a small molecule to an antibody’s surface is a process called antibody “labeling.” Labeling antibodies with small molecules such as biotin or fluorophores...

DNA gel electrophoresis is one of the most widely used analytical techniques in molecular biology, providing a simple and reliable way to separate DNA fragments...

Do you have a favorite restaurant that you love because you know exactly how great the experience is going to be? There are probably a...

Using the wrong agarose can lead to smearing or poor separation. The good news is that once you know how to evaluate agarose and choose...
