Deep Overview About RT-qPCR
by Adriana Gallego, Ph.D.

by Adriana Gallego, Ph.D.
Real-time quantitative reverse transcription PCR or qRT-PCR is a technique used in molecular biology that enables reliable detection and measurement of products generated during each cycle of a PCR process.
Among the qRT-PCR applications are:
However, to do qRT-PCR, it is important to talk about the basics of PCR and qPCR. This article will summarize the principles of PCR and qPCR, and the differences between RT-PCR and qRT-PCR.
Polymerase chain reaction, or PCR, is a basic molecular biology technique that has been used for decades and has been helping researchers open new scientific horizons.
The PCR technique consists of a reaction where a DNA fragment is copied over and over, exponentially.
The idea behind this is to amplify a DNA sequence to get several copies, which can be used in many downstream applications (low-abundance sequences amplification for cloning, pathogen, and polymorphisms detection, among others).
A PCR reaction contains:
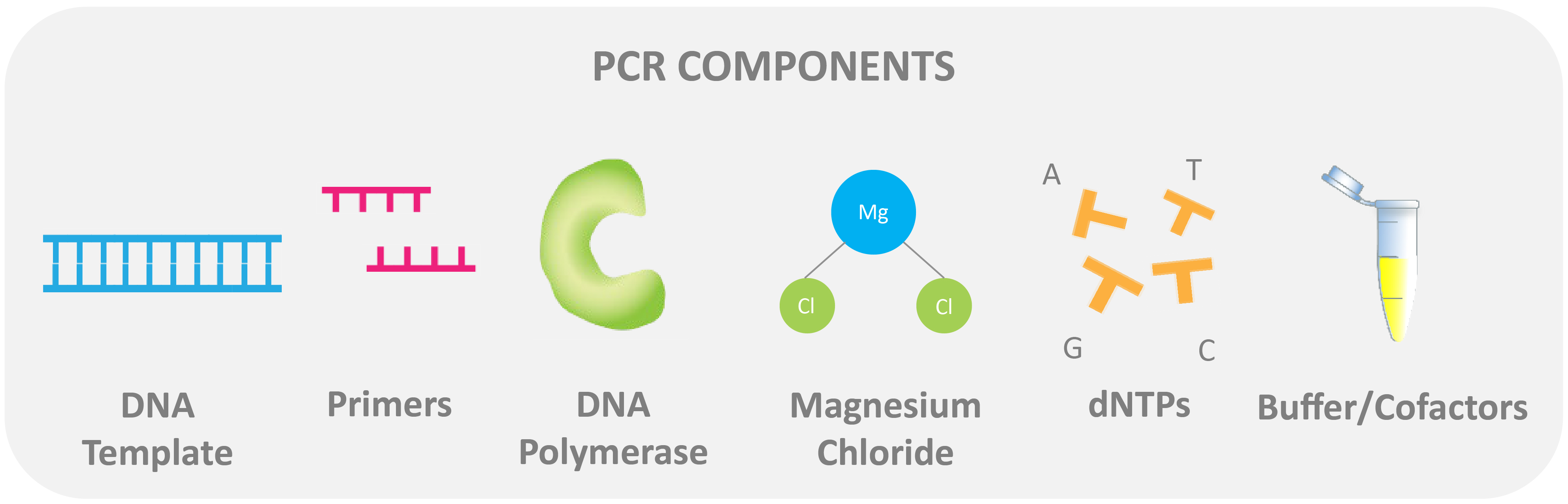
DNA template: this is the target sequence to start the reaction. For traditional PCR, the DNA is double-stranded.
Flanking oligonucleotide primers: Primers are joined at the ends of the DNA template sequence and are used to “identify” the target DNA within the sample.
Magnesium: This element is essential as a cofactor for DNA polymerase. Magnesium is added in the form of magnesium chloride to the master mix.
DNA polymerase: Usually, Taq polymerase is used. Taq polymerase is a thermostable enzyme and is used for polymerization (the process of adding nucleotides to the three prime (3')-end of a target DNA sequence).
Nucleotides: Nucleotides (pictured as dNTPs) are the building blocks of nucleic acids. Both DNA and RNA are composed of nucleotides.
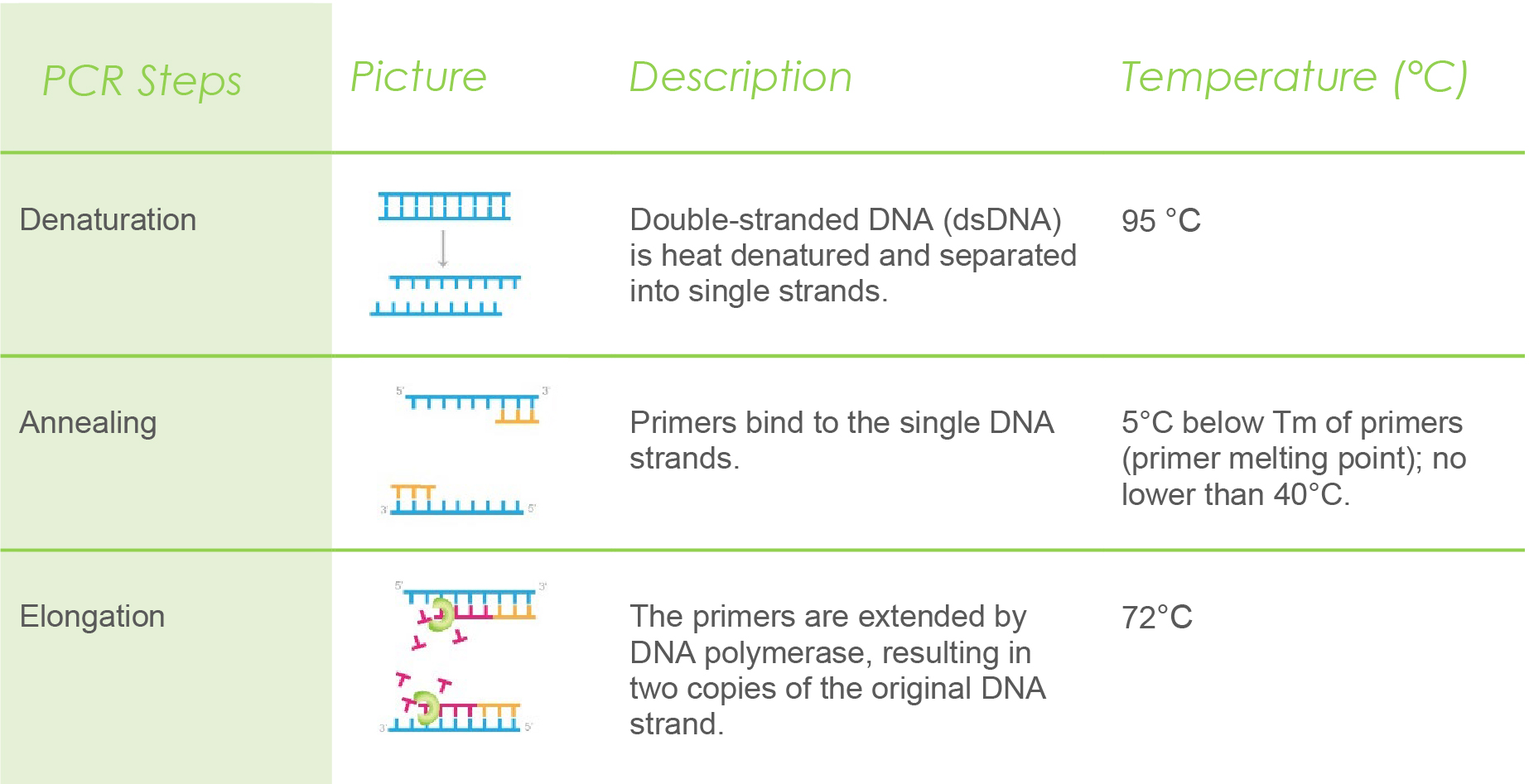
Polymerase chain reaction is carried out in cycles. Each cycle involves three steps: denaturation, annealing, and elongation. Generally, PCR can run up to 40 cycles to amplify the number of DNA copies.
Although quantitative PCR, or qPCR can help you answer similar questions as traditional PCR, qPCR, provides the ability to observe amplification in real-time, allowing you to quantify the number of copies in the starting material.
Quantification is essential to answer more profound questions about gene expression analysis and validate microarray and NGS results.
The qPCR technique differs from the traditional PCR in that the former introduces a probe (a designed oligonucleotide) or a fluorogenic dye to hybridize to the target sequence. Cleavage of the probe during PCR, because of the 5' nuclease activity of Taq polymerase, can be used to detect amplification of the target-specific product via fluorescent labeling.
The qPCR process has disadvantages and advantages compared to PCR. For instance, although qPCR is more expensive and requires specialized skills and equipment, qPCR allows you to visualize how your PCR proceeds, which facilitates the measurement of your DNA copies in real-time.

Now that you know the difference between PCR and qPCR, we can understand the concept of RT-qPCR.
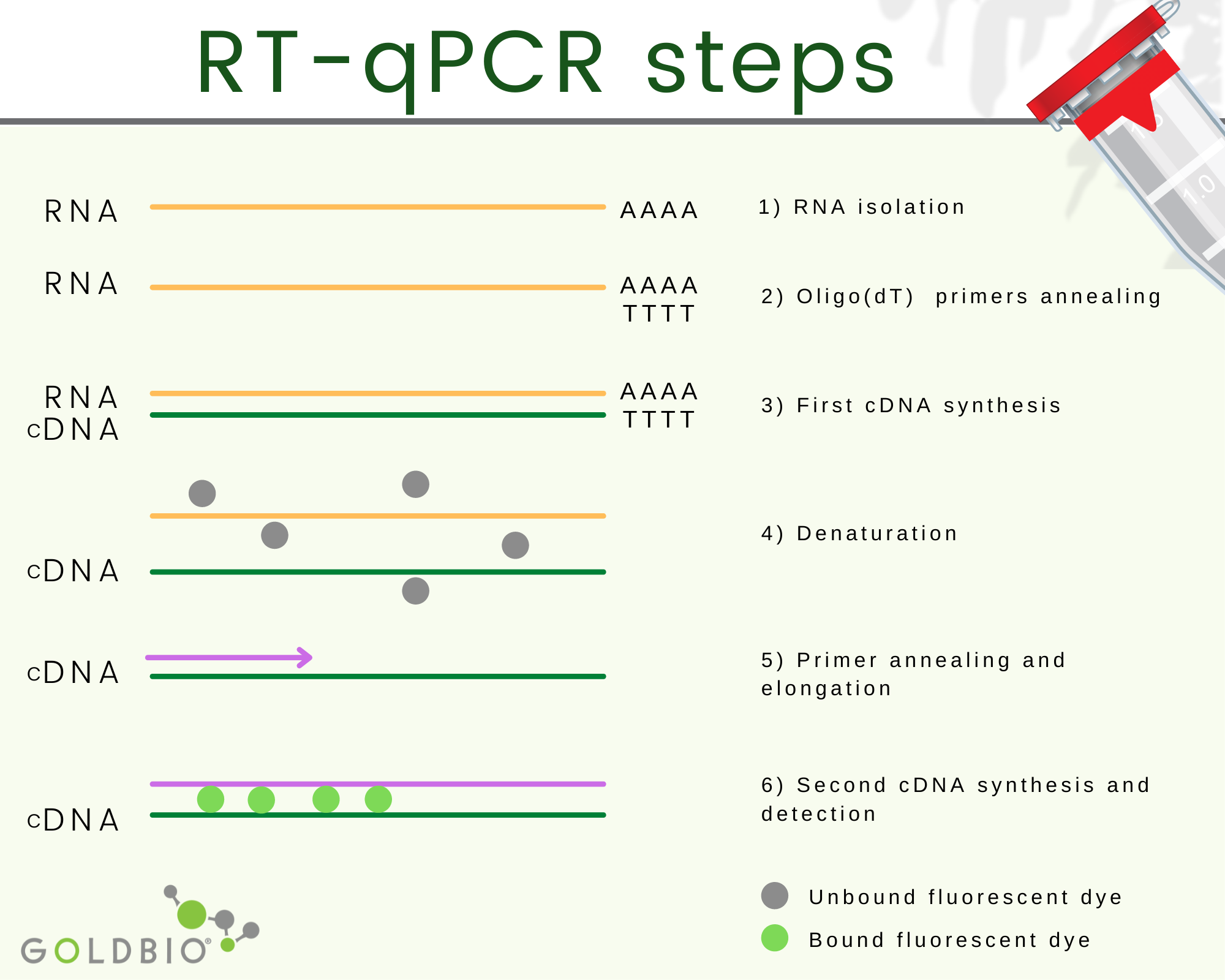
Quantitative reverse transcription PCR or RT-qPCR is a quantitative PCR where complementary DNA (cDNA), which is RNA transduced back into DNA, is used as starting material.
The complementary DNA or cDNA is the process where the RNA is transduced back to DNA.
Obtaining cDNA happens in two steps. First, a hybrid RNA/DNA is produced with the help of a reverse transcriptase enzyme. Second, a polymerase with the H function is used, which removes the RNA strand from the hybrid leaving the first DNA strand behind.
Afterward, a second DNA strand is synthetized using the first DNA strand template with the help of DNA polymerase. In this way, double-strand DNA is produced from RNA.
RNA is a very unstable molecule; therefore, it is tough to handle RNA for experimentation, and this includes PCR techniques.
Thanks to the reverse transcriptase enzyme (an RNA-dependent DNA polymerase), it is possible to synthesize DNA using RNA as the template. However, this also means there is an extra step involved when doing RT-qPCR.
Additionally, the fluorescent labeling by using reporter fluorogenic dyes (green like SYBR Green) enables researchers to collect data as PCR progresses.
SYBR Green exhibits slight fluorescence when in solution but emits a strong fluorescent signal upon binding to DNA.
Furthermore, ROX, also known as carboxyrhodamine, is an inert fluorescent dye that can be added to the qPCR master mix. The ROX fluorescent signal is stable throughout the RT-qPCR process, and it is not affected by the reaction. Consequently, it is used to normalize differences in signal intensity.


A His-tag is a stretch of 6-10 histidine amino acids in a row that is used for affinity purification, protein detection, and biochemical assays. His-tags...

Competent cells such as DH5a, DH10B, and BL21 will maintain their transformation efficiency for at least a year with proper storage. It is important to...

Ni2+ ions give nickel agarose beads their characteristic blue color. This blue color can fade or disappear completely when loading his-tagged proteins onto the column....

Nickel agarose beads change from blue to a brown or black color when the nickel ions have been reduced from a Ni2+ to a Ni1+...
