From Labs to Life: The Versatility of Purified Proteins in Practical Settings
by Simon Currie, Ph.D.

by Simon Currie, Ph.D.
Purified proteins help feed the world, keep us healthy, improve common products, and even begin solving pollution problems.
Purified proteins are proteins that have been isolated away from the many other types of molecules that they normally coexist with inside cells. And they are key ingredients in a range of products, technologies, and therapies.
Researchers use purified proteins to unravel biological mysteries, to investigate the causes of diseases, and figure out how to treat them. Nanoscale purified proteins can become medicines and important supplies in devices within the medical, food, environmental, and chemical industries.
In this article, we will take a brief look at each of these categories to understand their importance and learn about deeper areas within these categories that researchers are concentrating on.
Applications for purified proteins
Before we dive straight into the applications, let’s just take a moment to understand what protein purification is, and then define a few important terms.
Protein purification is a technique used in modern biochemistry and molecular biology. Traditionally, proteins were purified from cells, cell lines, and tissues that naturally have high expression of the protein of interest.
Examples include insulin from dog pancreas, respiratory chain proteins from cow heart, thrombin from cow blood, lectins from plant seeds, and botulinum neurotoxins (Botox®) from the bacteria Clostridium botulinum.
In modern times, proteins with research and commercial interest are often overexpressed in, and purified from, genetically engineered organisms.
These are called “recombinant” proteins because they are expressed by pieces of DNA that do not normally exist together in nature but have been recombined together by scientists.
Obtaining recombinant human proteins from bacteria, yeast, and plant cells led to products with more desirable traits that are also more reliably available and cheaper to produce.
Protein:
A protein is a biological polymer composed of amino acids.
Peptide:
A peptide is a short protein, typically less than 50 amino acids in length. In comparison, natural proteins are routinely hundreds, even up to a few thousand amino acids long.
Enzyme:
An enzyme is a general term for a protein that catalyzes a chemical reaction. In this article we will discuss many hydrolases, or enzymes that break down biological polymers using water, including proteases, lipases, and glycosidases.
Protease:
Proteases are a type of enzyme that cut proteins into smaller pieces. Different proteases have distinct specificities for the protein sequence that they cleave.

Figure 1. Schematic
of a protease cutting a protein into smaller pieces.
Lipase:
Lipases are enzymes that cut lipids by hydrolyzing triglycerides into fatty acids and glycerol.
Glycosidase:
Glycosidases are enzymes that cut long oligosaccharides into smaller sugar units. Examples include cellulases, amylases, and lactases, which each cut different types of carbohydrates.
Recombinant Protein:
A recombinant protein is obtained by using genetic engineering to express the protein in host cells from a different species. For example, recombinant human proteins are often obtained by expression in bacteria, yeast, or plant cells.
Directed evolution:
Directed evolution is the process wherein scientists evolve a new form of a protein in the lab to improve traits or introduce new functionalities. For example, a protein can be evolved to be more stable, or to improve activity under certain experimental conditions such as high temperature or acidic pH.
The cells in our bodies are the most complicated 4D jigsaw puzzles in the universe! They contain a complex mixture of billions of macromolecular pieces – proteins, lipids, and nucleic acids – that dynamically interact with each other to enable life as we know it.
Proteins carry out many of the biochemical activities that sustain life. In such a complicated environment, how do we sort out which molecules are important for which functions? As we will discuss below, purified proteins have played a key role in unraveling mysteries in human health and disease.
Purified proteins are also used in applications in a wide range of industries. Traditionally, these enzymes were sourced from naturally occurring ingredients such as:
However, many of these traditional sources are being replaced by, or supplemented with, recombinant forms of the key proteins in these processes. There are many advantages to using recombinant proteins:
Below, we will cover many of the different research and commercial applications that purified proteins are used in.
Purified proteins are key reagents that have helped scientists uncover fundamental biological discoveries.
One way to figure what individual proteins do is to purify them away from the complex cellular environment, and perform functional and structural studies on the protein in isolation. Examples of what was learned from such studies include:
Another way to figure out the functions of proteins is to observe the mutations that occur in diseases that affect humans and other animals.
For example, mutations in hemoglobin lead to its aggregation and causes sickle cell anemia. Causative protein mutations are known in numerous other diseases including cystic fibrosis, Duchenne muscular dystrophy, Huntington’s disease, Alzheimer’s disease, mad cow disease, prostate cancer, breast cancer, and Ewing’s sarcoma, just to name a few.
Purified proteins are also important in the drug discovery process and have led to the discovery of new treatments for chronic myeloid leukemia and prostate cancer, among other diseases.
Diabetics have trouble regulating their blood sugar, which over time leads to serious health problems such as vision loss, heart disease, and kidney disease.
Over a hundred years ago Frederick Banting, a surgeon from Toronto, envisioned a breakthrough treatment for diabetics. Banting realized that a number of pancreatic injuries did not cause diabetes. From these medical reports, he deduced that the Islet of Langerhans, a specialized part of the pancreas, must excrete a substance into the bloodstream to regulate sugar levels.
Within a year Banting had prepared an extract from dog pancreas that was effective at controlling blood sugar. In another year, with the help of biochemist James Collip, they isolated extract that was sufficiently pure for human use.
As a nod to the Islet of Langerhans, they called this purified extract “isletin” which later morphed into the current term insulin.
This rapid success transformed the treatment of diabetes for which Banting received the Nobel Prize in Physiology and Medicine.
Despite this transformative success, a substantial minority of diabetic patients developed an allergic reaction to insulin from dogs and other animals (Buse et al., 2021). Moreover, relying on animal pancreas as a source led to a limited and erratic supply chain for this crucial medicine.
It was several decades later before we understood that the active ingredient in purified pancreas secretions is a single protein. Additional decades and scientific advances enabled researchers to express human insulin in bacterial cells whereby the protein is extracted and purified from bacterial cells in a so-called recombinant form.
The use of human insulin drastically reduces the number of diabetic patients that develop an allergic reaction, and allows more reliable and affordable production of this crucial medicine that is used by over 150 million patients worldwide (Buse et al., 2021).
There are numerous other purified proteins that also play key roles in the medical field. These proteins are life-saving cancer medicines and treatments that help people lose weight.
Purified proteins are also important ingredients in medical diagnostics for Covid-19 and in waterproof adhesives that will serve as waterproof stitches in the future.
Indeed, proteins are transforming medical treatment in virtually every realm of care.
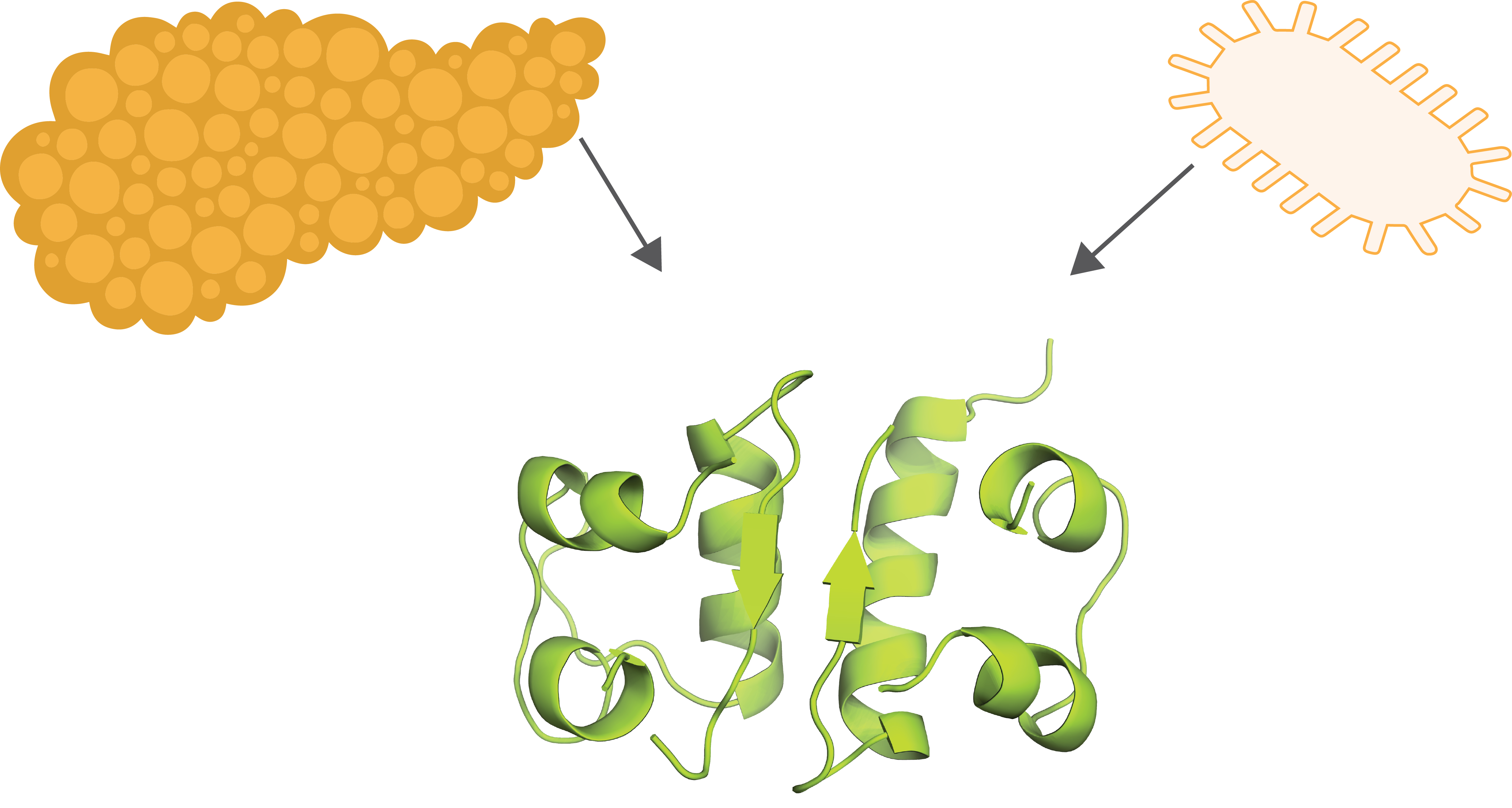
Figure 2. Insulin
(PDB:1BEN) was traditionally purified from animal pancreas but human
recombinant protein is now expressed and purified from bacteria (Smith et al.,
1996).
What do cheese, beer, cake, cookies, and bread all have in common? Enzymes are used to generate these delicious treats!
Traditionally these enzymes come from source ingredients and the microorganisms used to mature these products.
For example, alpha-amylase traditionally comes from the grain used to make beer and baked goods.

Figure 3. Alpha-amylase
cuts carbohydrates into simple sugar molecules (left). Structure of alpha
amylase (left, PDB: 1MXD).
However, adding extra alpha-amylase from outside sources can help make the baking and brewing process more consistent and reproducible, and can be used to confer new properties to the finished product.
For instance, adding extra alpha-amylase delays bread from going stale. Furthermore, distinct alpha-amylases from different species have unique activity profiles, such as what pH and temperature they are active at. Through extensive experimentation, brewers used different alpha-amylases to brew beer with distinct sweetness, body, and mouthfeel profiles.
In many ways, the industrial revolution was a chemical revolution where humans learned how to harness chemical energy from natural sources and use that energy to create new products that make our everyday lives more convenient and unrecognizable to our recent ancestors.
Yet, this industrial/chemical revolution led to environmental crises: slowly degrading plastic waste piling up in landfills and in ocean garbage patches, toxic spills, and limited/nonrenewable energy sources, to name a few.
Naturally occurring and newly evolved proteins are being used to detect and resolve such environmental problems.
For example, poly(ethylene terephthalate) (PET) hydrolases degrade PET plastics into smaller monomers that can then be recycled into new plastics. So, plastic is kept out of landfills or the ocean and new plastic products are made without making new plastic.
However, PET hydrolases have limited activity and stability that prevent them from being more widely used. Researchers have overcome this limitation by using directed evolution to generate a new PET hydrolase that is more active under a variety of experimental conditions and is more stable (Lu et al., 2022).
These evolved enzymes can thus recycle more plastic, and can themselves be reused, making the overall process more cost efficient. Similar protein innovations are solving many of the other pressing environmental issues mentioned above.

Figure 4. PET
hydrolase (left, PDB:6IJ6) and depolymerization of poly(ethylene terephthalate)
reaction that it catalyzes (right). Amino acids that were mutated to stabilize
and activate PET hydrolase are shown in sphere format.
Perhaps this morning you used toilet paper and applied skin-care products - after washing your hands, of course, then ran the dishwasher.
Cosmetics and cleaning products contain bioactive peptides and enzymes, respectively, and enzymes are used in the paper-making process.
Cleaning products contain many enzymes that cleave biological molecules: proteases that cut proteins, glycosidases that cut carbohydrates, and lipases that cut lipids or fats.
Many of these enzymes are engineered to provide better cleaning performance under less-than-ideal conditions.
One example is a protease that was engineered to be more active in the cold wash temperatures that are commonly used for laundry in Europe (Safdar et al., 2023; Zada et al., 2023).
Furthermore, using biodegradable enzymes reduces the need for toxic chemicals, making many of these cleaning products more environmentally friendly.
As you can see, purified proteins play key roles in human health and disease, and are used in a variety of important and everyday products. If you want a deeper dive into any of these application areas, you may want to check out our other articles that do just that!
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The Protein Data Bank. Nucleic acids research, 28(1), 235–242. https://doi.org/10.1093/nar/28.1.235
Berman, H., Henrick, K., & Nakamura, H. (2003). Announcing the worldwide Protein Data Bank. Nature structural biology, 10(12), 980. https://doi.org/10.1038/nsb1203-980
Buse, J. B., Davies, M. J., Frier, B. M., & Philis-Tsimikas, A. (2021). 100 years on: the impact of the discovery of insulin on clinical outcomes. BMJ Open Diabetes Res Care, 9(1). doi:10.1136/bmjdrc-2021-002373
Carpenter, D. (2015). Fun with Enzymes. doi: https://beerandbrewing.com/fun-with-enzymes/
Chen, C. D., Welsbie, D. S., Tran, C., Baek, S. H., Chen, R., Vessella, R., . . . Sawyers, C. L. (2004). Molecular determinants of resistance to antiandrogen therapy. Nat Med, 10(1), 33-39. doi:10.1038/nm972
Cleton-Jansen, A. M., Collins, N., Lakhani, S. R., Weissenbach, J., Devilee, P., Cornelisse, C. J., & Stratton, M. R. (1995). Loss of heterozygosity in sporadic breast tumours at the BRCA2 locus on chromosome 13q12-q13. British journal of cancer, 72(5), 1241–1244. https://doi.org/10.1038/bjc.1995.493
Collins, S. J., & Groudine, M. T. (1987). Chronic myelogenous leukemia: amplification of a rearranged c-abl oncogene in both chronic phase and blast crisis. Blood, 69(3), 893-898. doi:10.1182/blood.V69.3.893.893
Deininger, M. W. N., Goldman, J. M., Lydon, N., & Melo, J. V. (1997). The Tyrosine Kinase Inhibitor CGP57148B Selectively Inhibits the Growth of BCR-ABL–Positive Cells. Blood, 90(9), 3691-3698. doi:10.1182/
Ge, H., Martinez, E., Chiang, C. M., & Roeder, R. G. (1996). Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods in enzymology, 274, 57–71. https://doi.org/10.1016/s0076-6879(96)74008-9
Hegele, R. A., & Maltman, G. M. (2020). Insulin's centenary: the birth of an idea. Lancet Diabetes Endocrinol, 8(12), 971-977. doi:10.1016/S2213-8587(20)30337-5
Hill, A. F., Desbruslais, M., Joiner, S., Sidle, K. C., Gowland, I., Collinge, J., Doey, L. J., & Lantos, P. (1997). The same prion strain causes vCJD and BSE. Nature, 389(6650), 448–526. https://doi.org/10.1038/38925
Iannuzzi, M. C., Stern, R. C., Collins, F. S., Hon, C. T., Hidaka, N., Strong, T., Becker, L., Drumm, M. L., White, M. B., & Gerrard, B. (1991). Two frameshift mutations in the cystic fibrosis gene. American journal of human genetics, 48(2), 227–231.
Kao, L., Krstenansky, J., Mendell, J., Rammohan, K. W., & Gruenstein, E. (1988). Immunological identification of a high molecular weight protein as a candidate for the product of the Duchenne muscular dystrophy gene. Proceedings of the National Academy of Sciences of the United States of America, 85(12), 4491–4495. https://doi.org/10.1073/pnas.85.12.4491
Linden, A., Mayans, O., Meyer-Klaucke, W., Antranikian, G., & Wilmanns, M. (2003). Differential regulation of a hyperthermophilic alpha-amylase with a novel (Ca,Zn) two-metal center by zinc. The Journal of biological chemistry, 278(11), 9875–9884. https://doi.org/10.1074/jbc.M211339200
Lu, H., Diaz, D. J., Czarnecki, N. J., Zhu, C., Kim, W., Shroff, R., . . . Alper, H. S. (2022). Machine learning-aided engineering of hydrolases for PET depolymerization. Nature, 604(7907), 662-667. doi:10.1038/s41586-022-04599-z
Matthews, J. C., Hori, K., & Cormier, M. J. (1977). Purification and properties of Renilla reniformis luciferase. Biochemistry, 16(1), 85–91. https://doi.org/10.1021/bi00620a014
Miguel, A. S. M., da Costa Figueiredo, E.V., Lobo, B.W.P., Dellamora-Ortiz, G.M. (2013). Enzymes in Bakery: Current and Future Trends. In Food Industry.
Murphy, M. P., & LeVine, H., 3rd. (2010). Alzheimer's disease and the amyloid-beta peptide. J Alzheimers Dis, 19(1), 311-323. doi:10.3233/JAD-2010-1221
Nishio, I., Tanaka, T., Sun, S. T., Imanishi, Y., & Ohnishi, S. T. (1983). Hemoglobin aggregation in single red blood cells of sickle cell anemia. Science (New York, N.Y.), 220(4602), 1173–1175. https://doi.org/10.1126/science.6857241
Persichetti, F., Ambrose, C. M., Ge, P., McNeil, S. M., Srinidhi, J., Anderson, M. A., Jenkins, B., Barnes, G. T., Duyao, M. P., & Kanaley, L. (1995). Normal and expanded Huntington's disease gene alleles produce distinguishable proteins due to translation across the CAG repeat. Molecular medicine (Cambridge, Mass.), 1(4), 374–383.
Safdar, A., Ismail, F., & Imran, M. (2023). Characterization of Detergent-Compatible Lipases from Candida albicans and Acremonium sclerotigenum under Solid-State Fermentation. ACS Omega, 8(36), 32740-32751. doi:10.1021/acsomega.3c03644
Schneider, E., & Altendorf, K. (1985). All three subunits are required for the reconstitution of an active proton channel (F0) of Escherichia coli ATP synthase (F1F0). EMBO J, 4(2), 515-518. doi:10.1002/j.1460-2075.1985.tb03658.x
Smith, G. D., Ciszak, E., & Pangborn, W. (1996). A novel complex of a phenolic derivative with insulin: structural features related to the T-->R transition. Protein science : a publication of the Protein Society, 5(8), 1502–1511. https://doi.org/10.1002/pro.5560050806
Son, H. F., Joo, S., Seo, H., Sagong, H. Y., Lee, S. H., Hong, H., & Kim, K. J. (2020). Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzyme and microbial technology, 141, 109656. https://doi.org/10.1016/j.enzmictec.2020.109656
The PyMOL Molecular Graphics System, Version 2.5.2 Schrödinger, LLC.
Tran, C., Ouk, S., Clegg, N. J., Chen, Y., Watson, P. A., Arora, V., . . . Sawyers, C. L. (2009). Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science, 324(5928), 787-790. doi:10.1126/science.1168175
Walsh, G. (2005). Therapeutic insulins and their large-scale manufacture. Appl Microbiol Biotechnol, 67(2), 151-159. doi:10.1007/s00253-004-1809-x
Zada, S., Khan, M., Sajjad, W., Rafiq, M., Sajjad, W., & Su, Z. (2023). Isolation and characterization of a cold-active, detergent-stable protease from Serratia sp. TGS1. J Basic Microbiol, 63(10), 1165-1176. doi:10.1002/jobm.202300192

Maybe you are about to start working with D-luciferin, and you begin reviewing protocols. But what you’re doing is a little different. Or what you...

Handles are common in everyday life. We use them to help us open doors, cabinets, or to hold coffee mugs, cookware, and tools. They are...

Labeling antibodies with biotin or a fluorophore enables scientists to detect, track, and quantify that antibody and the molecules that it binds to. We cover...

Covalently conjugating a small molecule to an antibody’s surface is a process called antibody “labeling.” Labeling antibodies with small molecules such as biotin or fluorophores...
