E. coli Homologous Recombination: How It Works and Influences on Cloning
by Pallabi Roy Chakravarty, Ph.D.

by Pallabi Roy Chakravarty, Ph.D.
Homologous recombination is the exchange of sequences that occurs between two similar (homologous) nucleic acid molecules. This happens in both prokaryotes and eukaryotes. The process is well-characterized in the model lab bacterium E. coli, and it has implications for genetic engineering.
Homologous sequences are similar, but not identical. As an example, here is an illustration of two DNA molecules that are homologous.

If you note, the nucleotide sequences of molecules 1 and 2 differ only in two regions – highlighted with blue and pink boxes. This is an example of sequence similarity – they are mostly same with slight differences. And, from the perspective of molecular biology, molecules 1 and 2 are homologous.
Now, once homologous recombination occurs, the variation that occurs in one of the molecules, is represented here.
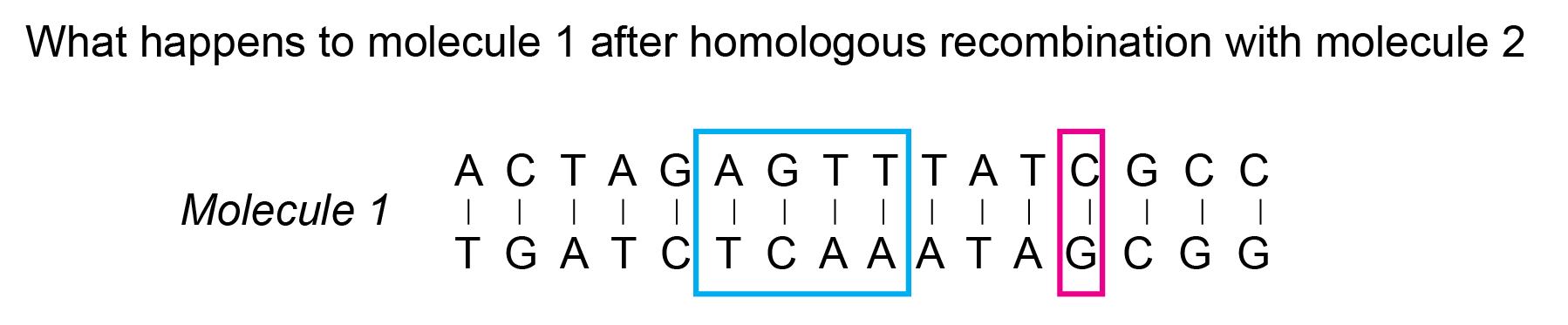
In this case, the variation is shown to be happening in the region highlighted in the blue box.
This is coincidental; the variation might have also occurred in the sequence in the pink box depending on how the pattern in which the recombination occurs between the two molecules.
In natural physiology, homologous recombination serves two main purposes.
First, to repair damage or breaks in DNA that can occur during DNA replication or be caused by UV light or other mutagens.
Second, to induce genetic variation in organisms by facilitating crossing over in eukaryotic chromosomes during meiosis and horizontal gene transfer in prokaryotes, such as bacteria.
Bacteria, including E. coli, have their own machinery for homologous recombination, and that will be the focus of this article.
However, the type of homologous recombination discussed in this article is not used as a tool for actually making genetic mutations or constructing recombinant DNA molecules.
There is another type of homologous recombination that is used as a tool for genetic engineering purposes.
For example, when you construct gene deletions or replace one DNA sequence with another in a cell’s genome or in vector DNA, this process is known as lambda Red recombineering. It utilizes proteins from the bacteriophage lambda, instead of E. coli’s own homologous recombination machinery.
Here is another thing to note – which will be a second aspect this article will cover: Because homologous recombination can induce variations in the DNA sequence, the cell’s underlying homologous recombination is turned off in E. coli cloning and expression strains.
This ensures unintended mutations/ variations do not occur in the cloned genetic construct or in the vector backbone.
Homologous recombination can be a challenging topic to understand. But its principles are important because once you understand the process, you will be able to better appreciate the value of its absence in cloning strains and what mutations turn off homologous recombination in cloning strains.
So, in this article, while discussing E.
coli
’s natural homologous recombination machinery, we will highlight more
about why it is
turned off in cloning and expression strains.
The process of E. coli homologous recombination
Importance of E. coli RecA mutants in cloning strains
E. coli has its own homologous recombination machinery to repair double-strand breaks in its genome and to produce genetic diversity through horizontal gene transfer.
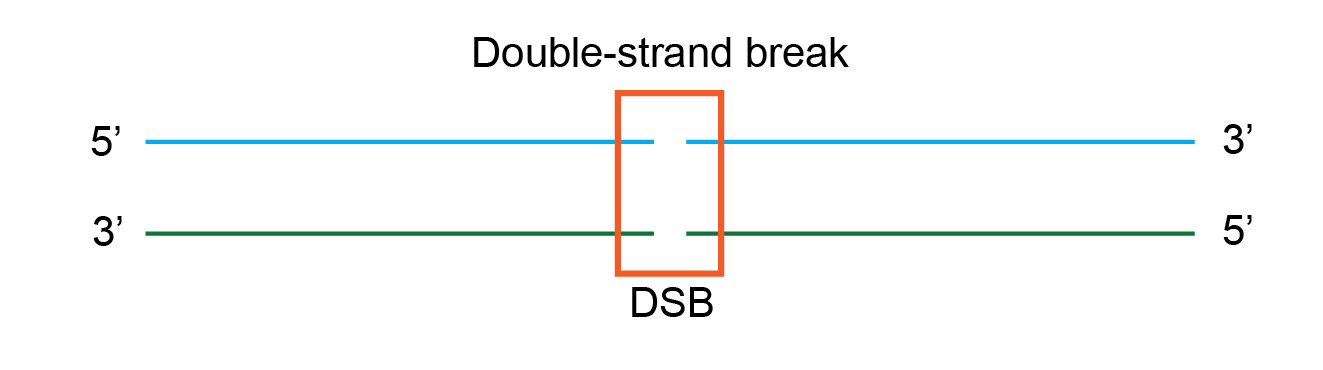
To illustrate the intrinsic process of homologous recombination in E. coli, here is an example of a DNA molecule that has a double-strand break.
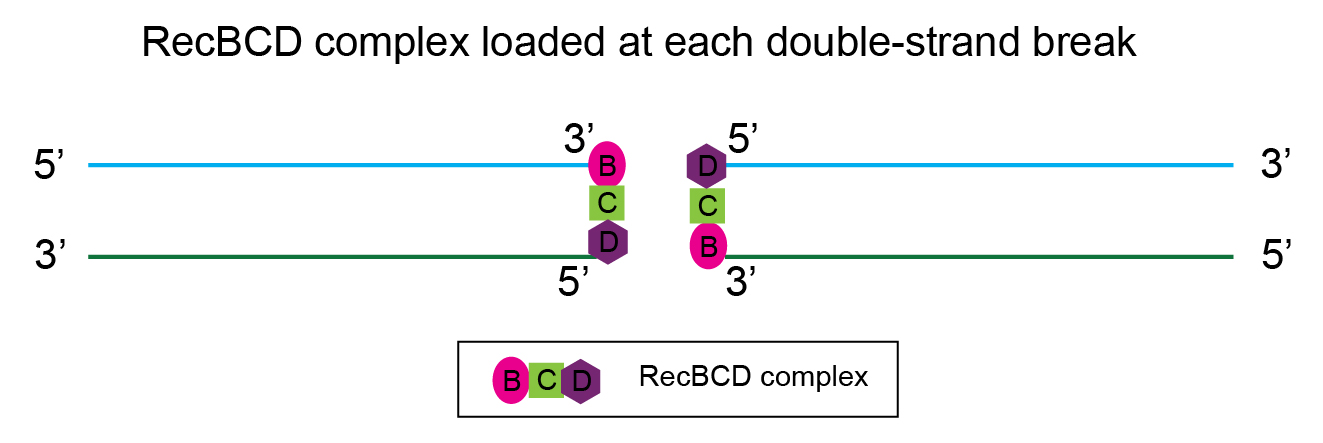
Next, the complex of three proteins RecBCD
assembles at each double-strand break.
RecB and RecD have helicase activity. They unwind the DNA duplex (double-strand). Additionally, the RecBCD complex has nuclease activity – the 5’ ends of the double-strand breaks are trimmed, producing 3’ overhangs. This is depicted in the illustration below. Technically, this is called strand resection.
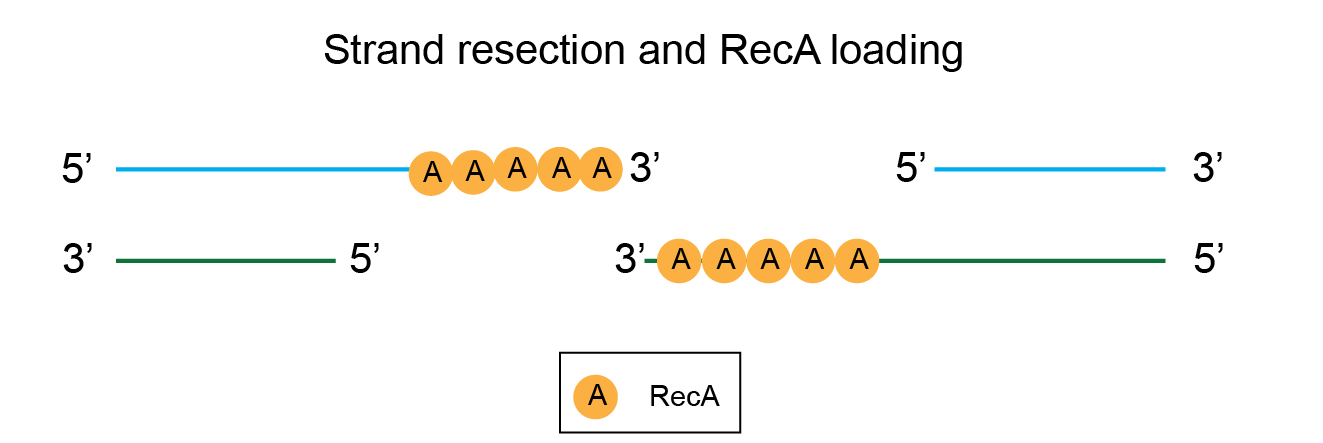
The 3’ overhangs are now loaded with the protein RecA, forming a nucleoprotein complex.
This RecA-coated single-stranded nucleoprotein searches for homologous DNA sequences within the cell. This is a very critical step in this entire process. Because of this, RecA mutant strains are not capable of homologous recombination.
While all Rec proteins are important for homologous recombination in E. coli, without RecA, homologous recombination is completely absent. Keeping this is mind, to avoid unintended mutations during genetic engineering, most E. coli cloning strains used in cloning are RecA mutants.
Once another DNA molecule is found that contains a homologous sequence, the RecA -covered nucleoprotein invades that intact DNA duplex. Using its sequence complementarity (because of homology between the two strands), it forms a triplex by binding in the major groove of the homologous duplex DNA.
As shown below, this triplex, with the displaced strand, looks somewhat like the letter D – and is called the D-loop.
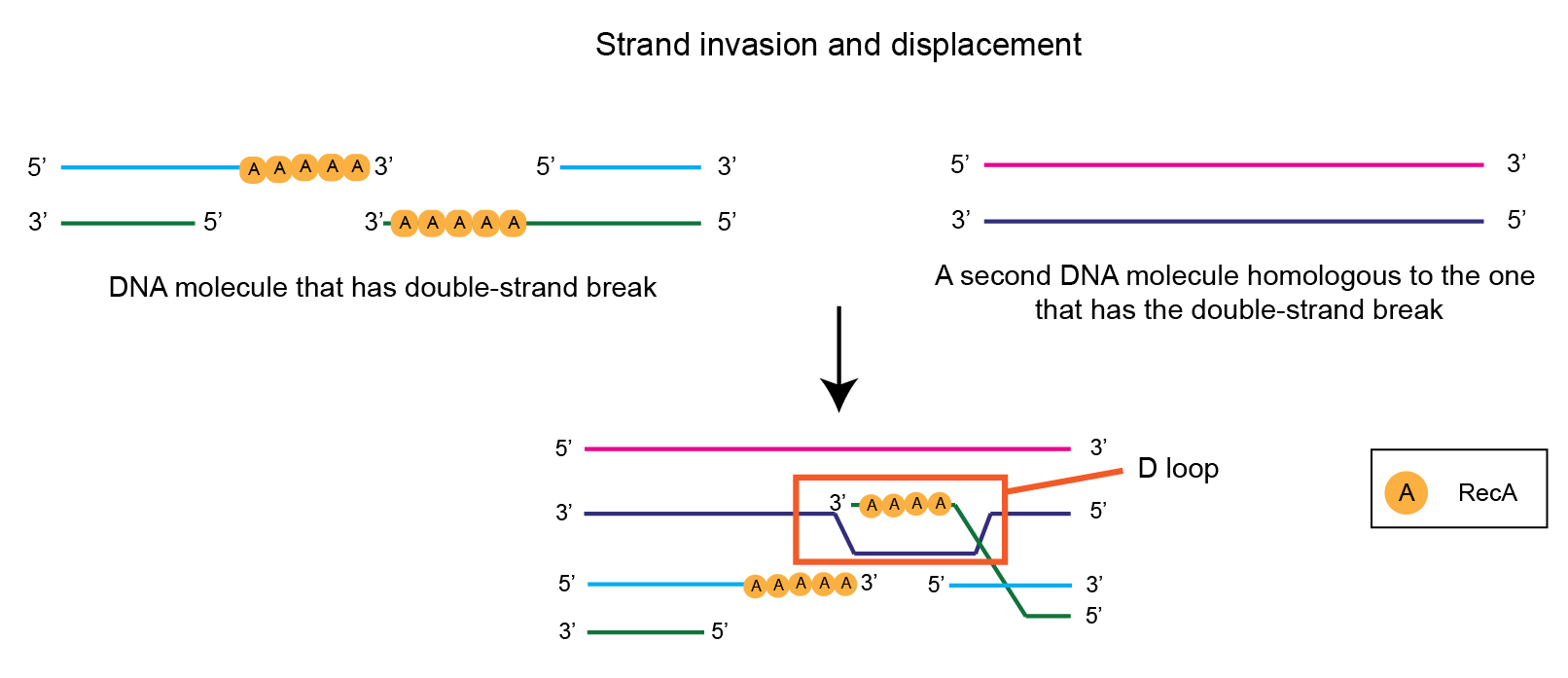
After this, the 3’ end of the invading strand is extended by DNA Polymerase I.
This makes the D-loop larger and larger. Ultimately, the D-loop becomes large enough that the displaced strand anneals to the other 3’ overhang (also coated with RecA) of the double-strand break.
This is depicted below.
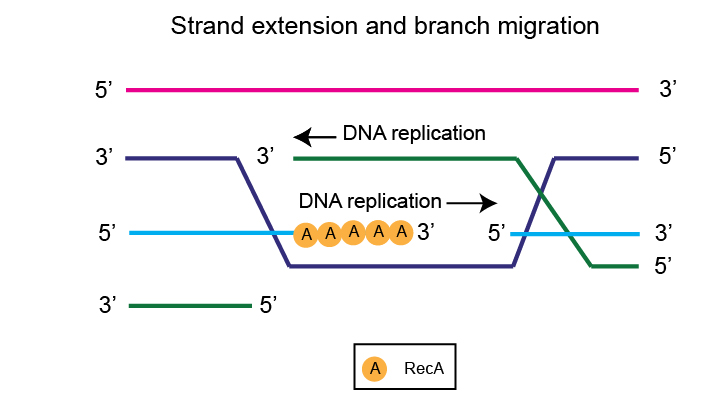
This 3’ overhang is also replicated and extended by DNA Polymerase I.
Ultimately, this span of recombination between the two homologous DNA molecules keeps extending. This is technically called branch migration.
Branch migration is facilitated by two proteins, RuvA and RuvB that leads to a second crossover between the two homologous DNA molecules, forming two heteroduplexes.
The points where these heteroduplexes are joined are called Holliday junctions.
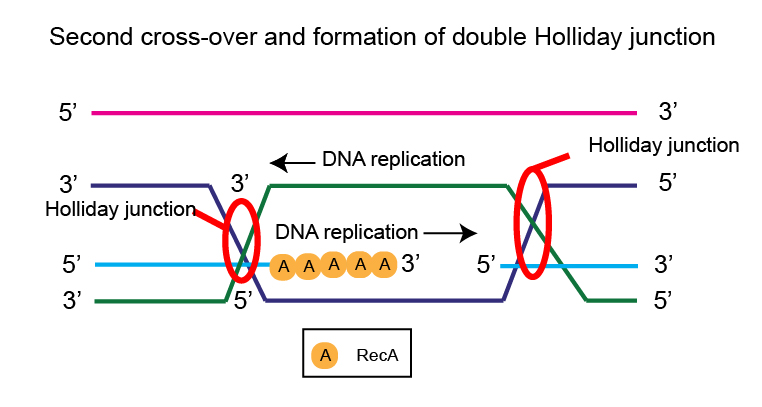
These Holliday junctions are resolved by the RuvC protein that has endonuclease activity and a DNA ligase, separated back into two different double-stranded DNA molecules.
In brief, this is how homologous recombination takes place in E. coli, using its own proteins (RecA, RecBCD, RuvABC etc.).
This process is utilized by E. coli (and other bacteria) not only to repair double-strand DNA breaks, but also to produce genetic diversity in the organism by recombining parts of its genome with other homologous DNA that it takes up from the surrounding environment through horizontal gene transfer (transformation, transduction and conjugation).
This is beneficial for the organism from an evolutionary diversity point of view – the higher the genetic diversity in an organism, the better off it is.
However, when you are using that
strain for cloning purposes, this might have serious implications.
And you need to be very careful of that. In the next section, we will have a
quick look into that, and the way around it.
Fun fact: Homologous recombination plays a role in human health. Just as it repairs double-stranded breaks in prokaryotes, eukaryotic cells, including human cells employ homologous recombination for DNA repair. The homolog of RecA in mammalian cells is the protein Rad51. So, Rad51 and other double-strand repair proteins have a huge role in malignancies.
Cloning strains like DH5α and DH10B do not have the equipment for homologous recombination due to the RecA mutation. And this is very necessary when using these strains as cloning hosts because it prevents random mutations in the cloned vector and insert.
This is illustrated in the figure below.
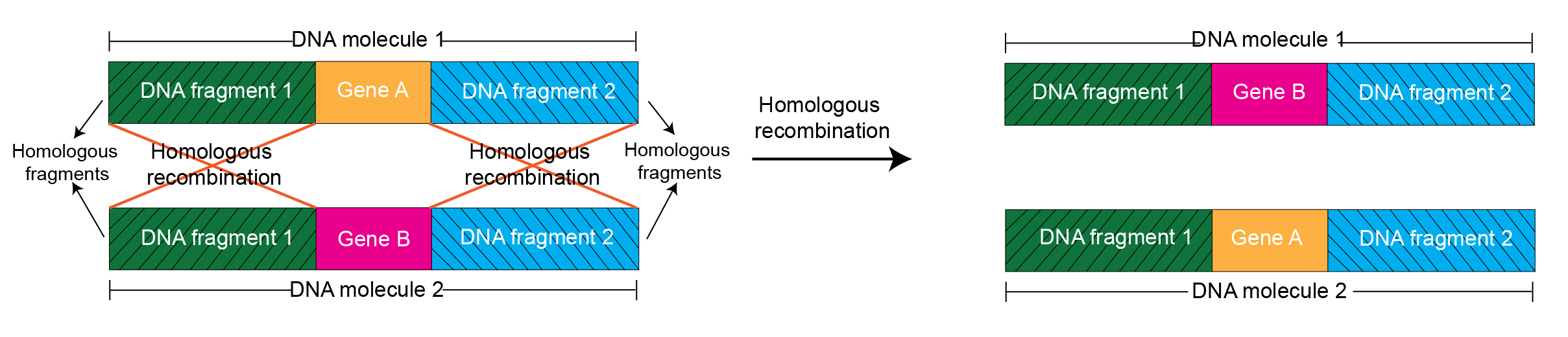
This illustration represents unwanted homologous recombination between two DNA molecules - the vector with the insert cloned in it and the host strain's genome.
This might occur unless the host strain's own homologous machinery is not blocked by mutating RecA.
For the two DNA molecules shown here, there are sequence homologies. The flanking regions are homologous, and the sequence in the middle – gene A and gene B in molecules 1 and 2 respectively, are not homologous.
In this type of a DNA sequence arrangement, homologous recombination can occur between the two molecules. This leads to swapping the nonhomologous sequences in the middle, as shown in the figure.
Now, in the context of a cloning experiment, consider DNA molecule 1 as a plasmid with gene A cloned in it. And molecule 2 is the bacterial genome.
Because of the unintended homologous recombination event, as shown in the figure, gene A that you painstakingly cloned in the plasmid has gotten changed to gene B. This would ruin your entire experiment.
To avoid such unintended mutations during cloning procedures, the cloning strain’s homologous recombination capability is blocked by making a mutation in RecA.
As a result, the sequence fidelity of your vector and insert is maintained during the cloning process.
Bell and Kowalczykowski. 2016. RecA: Regulation and Mechanism of a Molecular Search Engine. Trends Bichem. 41(6):491–507. doi:10.1016/j.tibs.2016.04.002
Dillingham and Kowalczykowski. 2008. RecBCD Enzyme and the Repair of Double-Stranded DNA Breaks. Microbiol Mol Biol Rev. 72(4): 642–671. doi: 10.1128/MMBR.00020-08
Douglas Wright et al. 2018. Homologous recombination and the repair of DNA double-strand breaks. JBC Thematic minireview. doi 10.1074/jbc.TM118.000372
Thomason et al. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr Protoc Mol Biol. Chapter 1: Unit 1.16. doi:10.1002/0471142727.mb0116s78
Thomason et al. 2023. Recombineering: Genetic Engineering in Escherichia coli Using Homologous Recombination. Current protocols. https://doi.org/10.1002/cpz1.656
West SC (June 2003). "Molecular views of recombination proteins and their control". Nature Reviews Molecular Cell Biology. 4 (6): 435–45.
Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R (2003). Molecular Biology of the Gene (5th ed.). Pearson/Benjamin Cummings.

IPTG and auto-induction are two ways to induce protein expression in bacteria. They work similarly, but have different trade-offs in terms of convenience. While IPTG...

The final concentration of IPTG used for induction varies from 0.1 to 1.0 mM, with 0.5 or 1.0 mM most frequently used. For proteins with...

A His-tag is a stretch of 6-10 histidine amino acids in a row that is used for affinity purification, protein detection, and biochemical assays. His-tags...

Competent cells such as DH5a, DH10B, and BL21 will maintain their transformation efficiency for at least a year with proper storage. It is important to...
