Types of PCR used for Genetic Research: Applications where different types of PCR play a vital role
by Katharine Martin

by Katharine Martin
Genetic research has exploded in recent decades with emerging technologies, breakthroughs in sequencing and the advancing sophistication of PCR. This brief overview examines a few applications or areas of genetic research and how PCR is used in these types of research.
As the field of genetic research has branched out, so too has PCR. Tailored variations of PCR have now been developed and employed to validate research, to be a primary tool for search or for up and downstream analysis.
What is genotyping: Genotyping uses sequencing information to determine genetic differences or variants in individuals or in biological populations. This type of technique is used to investigate a predetermined and very specific region of the genome. For example, when thinking about human DNA, a high percentage of it is identical from one individual to another (more than 99%), but there is still variation from one person to another which makes us very unique. Genotyping can be used to analyze some of those differences.
There is sometimes question about what the difference is between genotyping and sequencing. Sequencing looks at genomic information from beginning to end, whereas genotyping is looking at a very specific aspect of the genome and comparing it against known sequencing data. This comparison enables researchers to examine differences in the genetic makeup of an individual or population. Often, a book analogy is used to describe the difference between genotyping and sequencing where the whole book would be your genome and sequencing that book would be like reading all of the letters within the book. If there was a print error on some of the books, where a single letter on a specific page was missing, genotyping would be the tool examining that type of situation.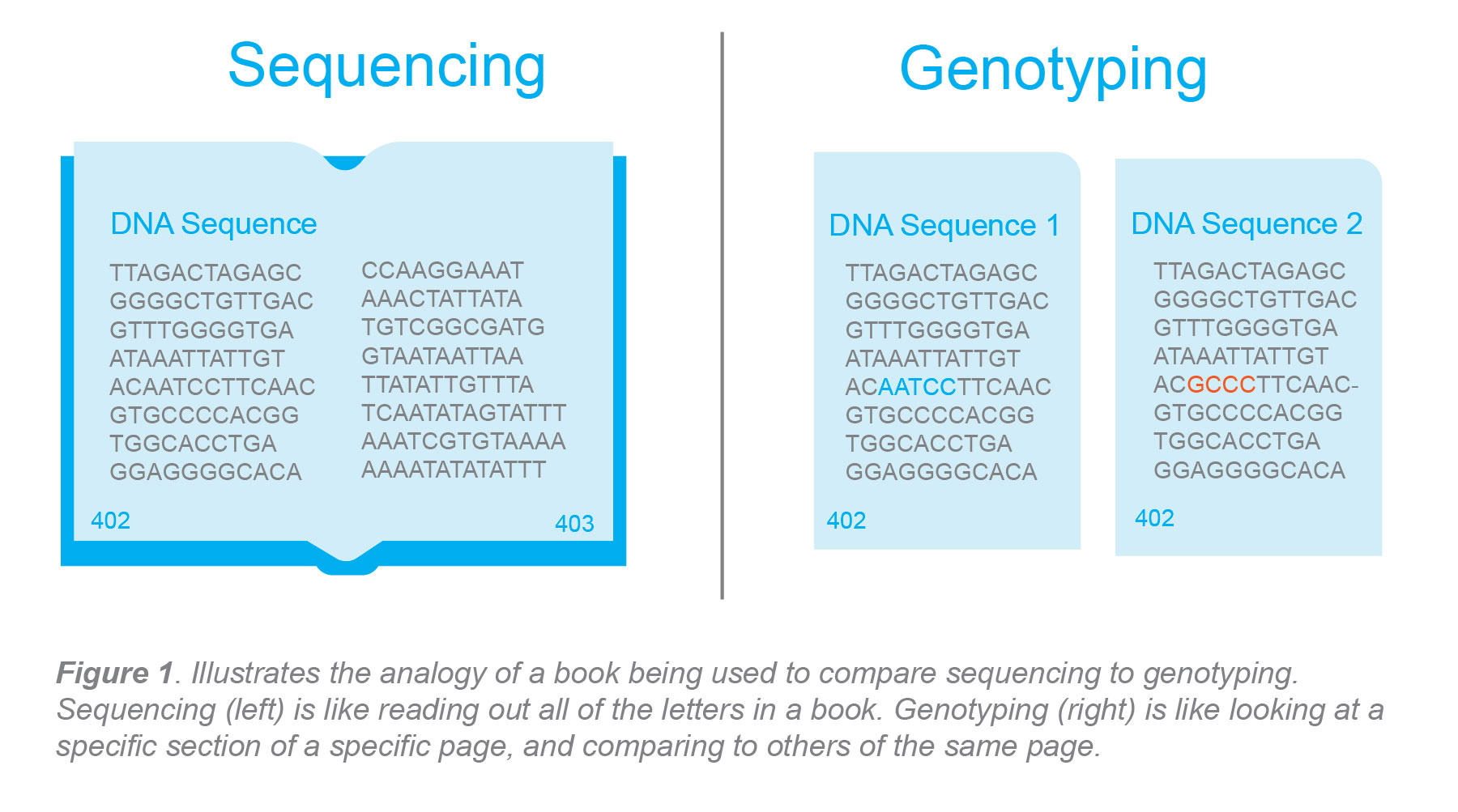
How is PCR used for genotyping: Understanding genetic makeup allows researchers more information about phenotypic traits. Quantitative PCR (qPCR) is only one type of technique used for genotyping and is used to identify single nucleotide polymorphisms (SNPs). Benefits to using qPCR is the ability to quickly and accurately get results, and minimizes the use of other hazardous material.
PCR for genotyping usually involves designing primers specific to the mutation or allele being studied.
Types of PCR used for genotyping:
What is microarray analysis: Microarray analysis looks at what genes are turned on during certain times with or without treatment (experimental conditions), and it can simultaneously evaluate thousands of samples.
For example, certain genes are switched on at night, which leads to the expression of proteins specific to that period of time. During the day, those night genes are switched off, and other sets of genes might be switched on, generating other types of proteins needed during daytime.
Microarray analysis is one way researchers can gain an understanding of what genes are turned on or off, and when.
In microarray analysis, a sample of tissue might be compared to a control sample in order to determine the differences in expression level between the two. During microarray analysis, a fluorescent dye is attached to small fragments of cDNA previously generated from the experimental and control samples. Red dye is used to label experimental cDNA and green dye is used to label control cDNA. The process takes place on a chip that has thousands of complementary DNA fragments to both the experimental and control. The mixture of fluorescently labeled control and experimental cDNA fragments are applied to the chip and hybridize to its complementary strand.
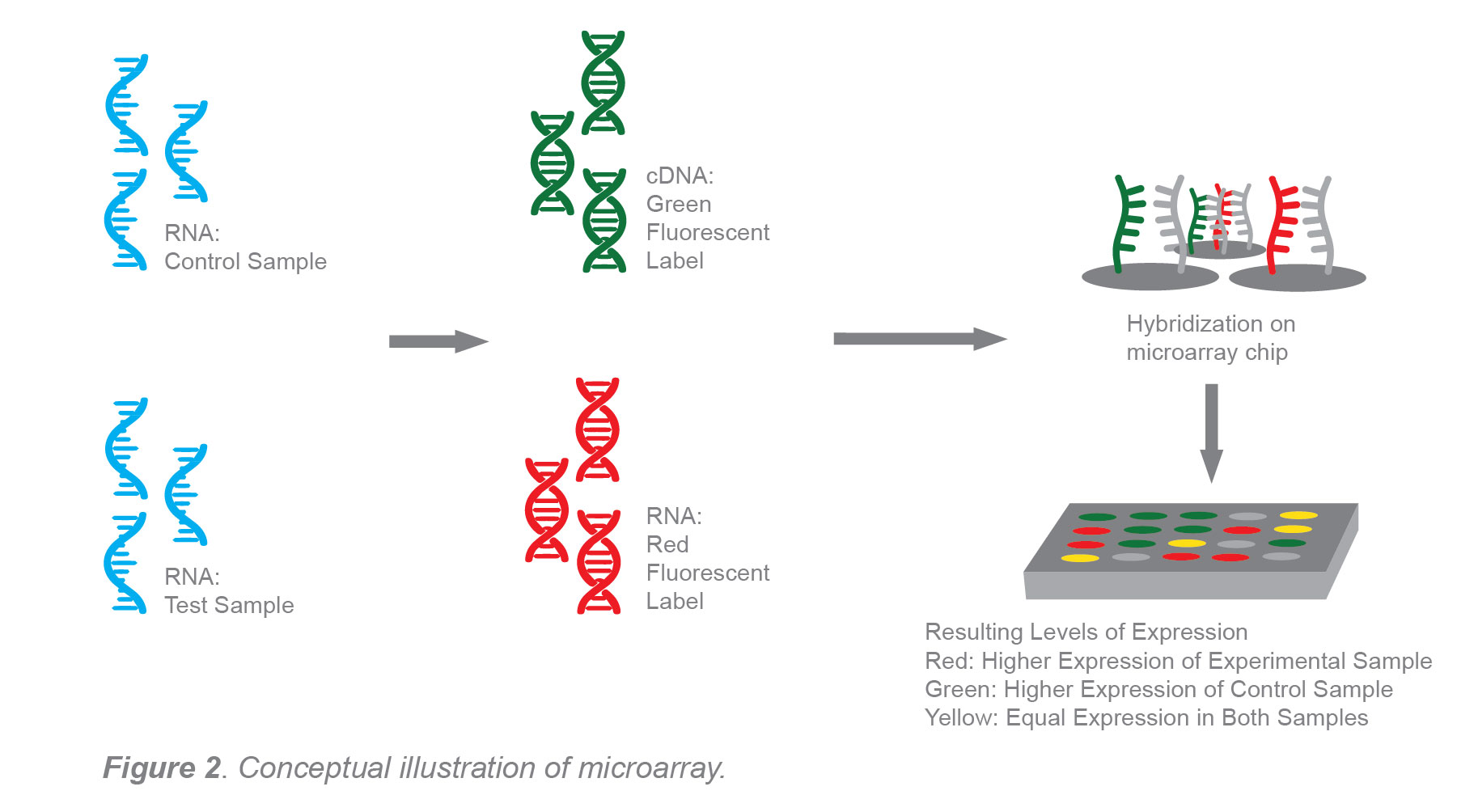
Results are color coded green, red or yellow. Green would represent high expression levels from the control, red would represent higher levels in the experimental sample, and yellow would represent equal expression levels.
How is PCR used for microarray: PCR can play different roles with respect to microarray analysis. RT-PCR is sometimes used prior to microarray to amplify cDNA for analysis. Quantitative PCR (qPCR), is an important validation step for microarray, allowing researchers to be certain of gene expression results. While microarray looks at relative expression levels from one sample to its comparative benchmark sample, qPCR provides quantitative expression levels.
Sometimes one method is chosen instead of another – a qPCR vs. microarray situation. If it comes down to choosing between methods, researchers should choose qPCR when evaluating gene expression in known samples. Microarray, on the other hand, is suited for situations where a larger number of genes are being studied, or for whole transcriptome analysis between two samples. In addition, microarray should be performed only when a good reference sequence is available.
The takeaway is that qPCR and microarray can go hand-in-hand, be used in place of one another, or be used for validation. Ultimately, the relationship between these two techniques will depend on experimental goals.
Types of PCR used for microarray:
What is RNA sequencing (RNA-seq): RNA-seq is a profiling method used to determine the types of genes that are switched on and at what level they’re being expressed. The process uses next generation sequencing (NGS) technology to analyze expression levels and quantity.
RNA-seq allows researchers to study total RNA makeup within a cell (transcriptome) and connect genomic information to protein functionality. Like microarray, RNA-seq gives us a glimpse into what genes are turned on during specific periods or under certain conditions, allowing us to compare protein production from different experimental conditions. Using NGS technology, researchers are now able to perform accurate RNA sequencing at high-throughput levels.
How is PCR used for RNA-seq: PCR can be used in RNA-seq library production (producing cDNA from mRNA) as well as amplification of samples for sequencing. The first part of the process involves reverse transcription in order to get cDNA. This is followed by amplification through PCR.
Emulsion PCR (ePCR) is another variation of PCR typically used for amplification prior to NGS. This type of PCR uses bead surfaces, water and oil. Emulsion PCR allows simultaneous amplification of each sequence without risk of contamination. Here, each bead acts as a microreactor for PCR, each containing one strand of DNA.

Bridge PCR is another method used to amplify sequences prior to NGS. Here, two types of oligos are fixed to a flow cell. Each oligo is complimentary to each adaptor flanking the DNA fragment. The flanking adaptors allow a bridge to form between the two types of oligos. After each copy is denatured, the single strands bridge to the oligos and the process gets repeated.

Types of PCR used for RNA-seq:
What is Single Nucleotide Polymorphisms (SNPs): Single nucleotide polymorphisms (SNPs) are single nucleotide substitutions within a certain region of a genome occurring in more than 1% of a given population. The human genome is about 99.5% identical from one individual to another. Of that 0.5% variation, SNPs contribute to about 90%. These variations are what make organisms unique. In humans, SNPs (pronounced snips) are the most common type of genetic variation and occur at a frequency of about one out of every 100-300 nucleotides. They are usually the result of copy or repair error, but can also be a result of diseases that cause mutations.
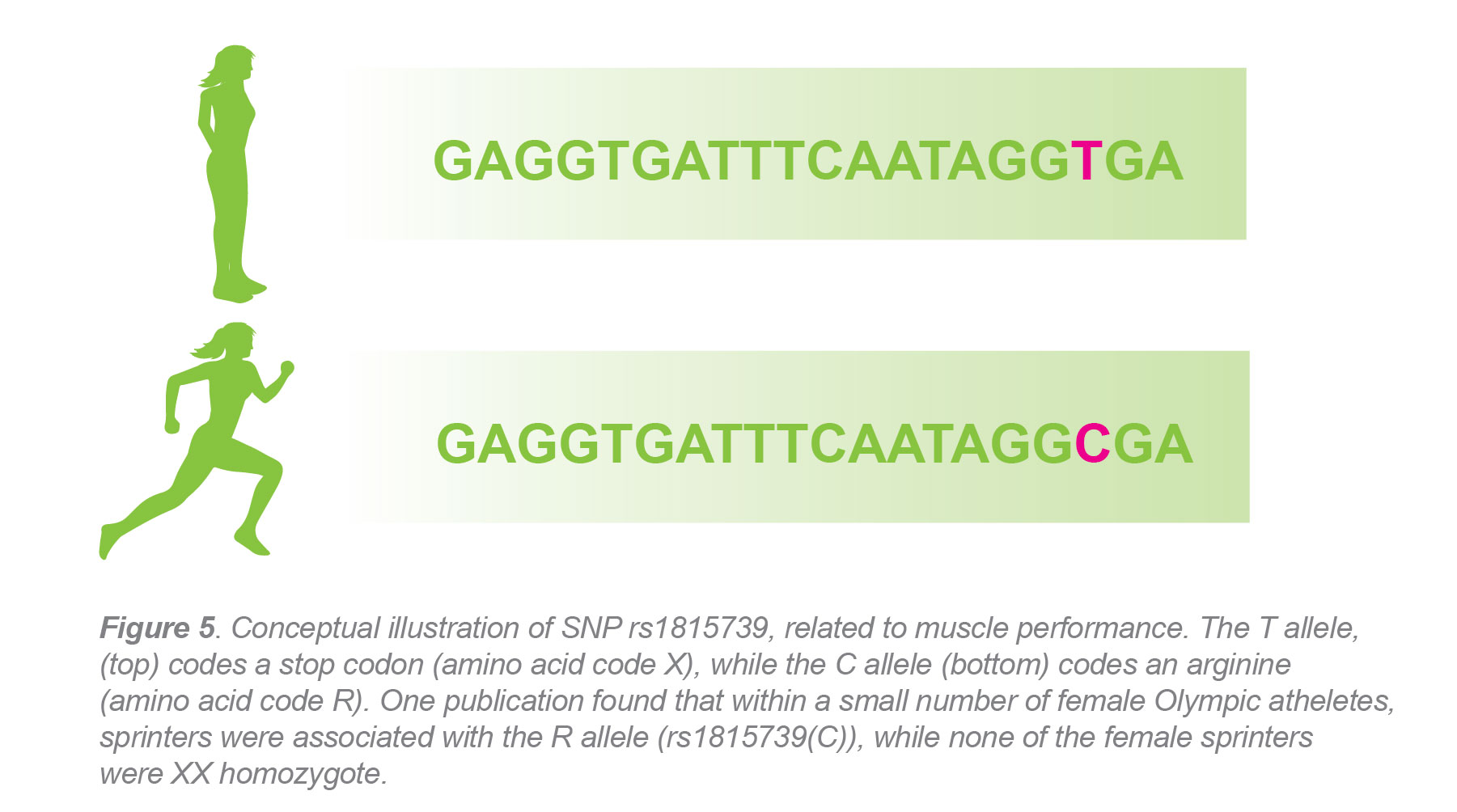
While SNPs occur quite often in the human genome, it will unlikely have an impact on cellular function unless the substitution occurs in a protein-coding region of the genome. Even still, SNPs that are found in non-coding regions can still lead to risk factors for diseases and cancer. SNPs are not only associated with disease phenotypes, but can also be associated with different characteristics in organisms. SNPedia is a wiki type database dedicated to studying variation in human genetics. Among some of the more popularly researched SNPs listed are variations causing characteristics in earwax, risk factors for breast cancer and type-2 diabetes, oxytocin receptor influences, and more.
Studying SNPs gives researchers insights into diseases, not just their cause – SNPs also help researchers identify regions relevant for specific diseases.
How is PCR used for analyzing SNPs: Recall that genotyping is the process of comparing DNA sequence information to identify differences in individuals and populations. Quantitative PCR (qPCR) is generally used for SNP genotyping in order to amplify and identify SNPs. Quantitative PCR will be ideal in situations where few SNPs are being studied. It’s suited for a large or small number of samples and allows for a quick and accurate analysis.
Amplification refractory mutation system PCR (ARMS PCR) is another type of PCR technique used to genotype SNPs. Here, sequence-specific primers for both the normal and the SNP-containing allele (mutant) are used in a single PCR reaction. The primers will amplify either the normal allele or the mutant allele, which can be visualized when analyzing the amplified fragments with agarose gel electrophoresis.
Other SNP genotyping techniques include sequencing and microarray analysis.
PCR types:
What is allele-specific expression analysis: Allele-Specific Expression (ASE) Analysis is a tool used to evaluate the difference in allelic expression between both parental alleles. The level of allelic expression within diploids will differ between the two inherited parental alleles, one being preferentially expressed over another. The regulation of allelic expression, which is unequally expressed, is called allele-specific expression (or allelic imbalance).
It’s thought that such expression differences are partly attributable to the cis-regulatory element, an area of noncoding DNA that regulates nearby gene transcription on the same strand of DNA. Examples of cis-regulator elements include regulatory regions, gene promoters, the terminator, the RNA ribosomal binding site and the enhancer.
Studying allelic expression provides more information on organismal phenotypic variation and diseases within populations.
What is gene expression analysis: Gene expression analysis evaluates gene regulation and gene products using RNA in order to determine patterns of gene expression. Gene expression research is important for understanding cell functionality through protein products, understanding overall activity, understanding the flow and regulation of information from DNA to RNA, understanding how the number of proteins produced impacts a system, etc.
How is PCR used for allele-specific expression and gene expression analysis: RNA sequencing (RNA-seq) is a common way to study gene and allele expression. RNA-seq can quantitatively show differences in expression levels among compared samples. However, PCR is another method researchers use to study expression.
Quantitative PCR (qPCR) and reverse transcription-quantitative PCR (RT-qPCR) are the types of PCR primarily used in gene expression research. One of the benefits of RT-qPCR is that it is suitable for more sensitive RNA quantification. It is also usually a better choice when analyzing only a few genes with a known sequence.
Types of PCR used to study allele-specific expression & gene expression:
What is DNA methylation and methylation analysis: The process in which a methyl group (CH3) is added to DNA is called DNA methylation. Methylation helps regulate gene expression by repressing transcription. This activity changes genetic function without altering DNA sequences and is one of many epigenetic mechanisms.
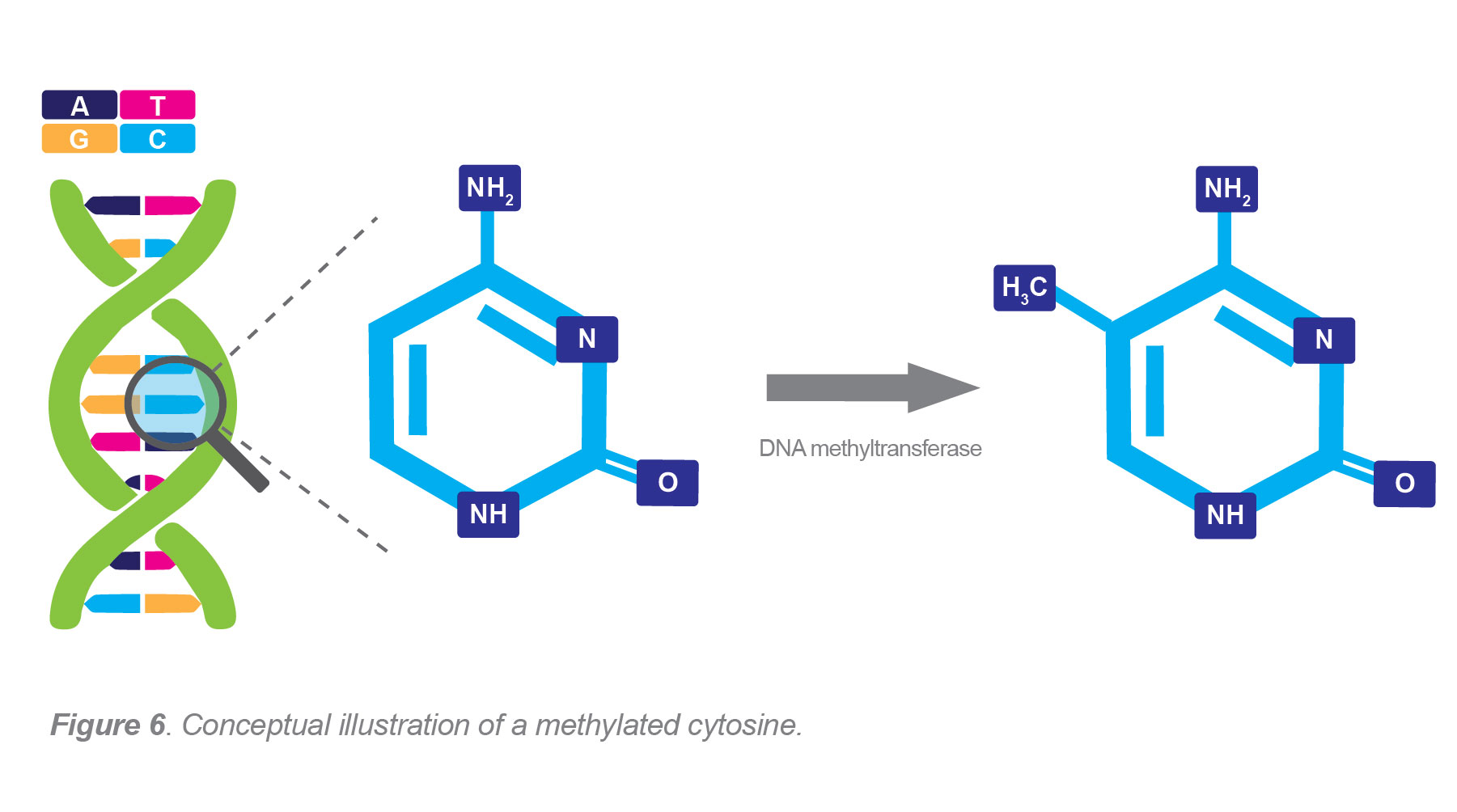
Gene expression processes attributed to DNA methylation include genomic imprinting, X-chromosome inactivation, development and aging.
Analysis of DNA methylation involves a broad set of methods used to address questions posed during research. Choosing a method for analysis will depend on several factors including goals, sample quality, needed sensitivity, budget, etc.
Sergey Kurdyukov and Martyn Bullock published “DNA Methylation Analysis: Choosing the Right Method” in 2016 with detailed guidance on how to choose an approach for analysis.
How is PCR used to analyze DNA methylation: Several types of PCR techniques exist to amplify and further study methylation. Different PCR types address certain obstacles or research needs.
Typically, PCR begins first with bisulfite conversion because downstream sequencing methods can’t distinguish between methylated cytosine and unmethylated cytosine. Bisulfite deaminates cytosine into a uracil. After conversion, DNA can be amplified by PCR, which enables further cloning and sequencing in order to further understand methylation.
The tricky part with bisulfite conversion is that DNA started with 4 nucleotides (A, T, C, G) and now only has 3 (A,T,G). This loss of DNA complexity also creates challenges to primer design.
Nested PCR is one way of reducing these challenges. Researchers have also employed other types of PCR techniques to overcome obstacles or satisfy the needs of their research. Below, in the types of PCR section are brief descriptions of each type of PCR.
Types of PCR used for DNA methylation analysis:
COLD-PCR (co-amplification at lower denaturation temperature-PCR) – COLD-PCR enables researchers to preferentially amplify minority alleles while in the presence of wild type alleles.
Here's the missing sticker you're looking for. You may find this sticker at vendor shows, in orders, or you can request a free 3-pack of randomly selected stickers for your collection. Be sure to share your collection using #GoldBioStickers.
(2014). Retrieved July 12, 2019, from https://www.nature.com/scitable/topicpage/gene-expression-14121669
23 and Me. (2019). What is the difference between genotyping and sequencing? Retrieved July 12, 2019, from https://customercare.23andme.com/hc/en-us/articles/202904600-What-is-the-difference-between-genotyping-and-sequencing-
23andMe. (n.d.). What are SNPs? Retrieved July 12, 2019, from https://www.23andme.com/gen101/snps/
Ababon, M. (2016, July 09). Get that Genotyping PCR to Work EVERY TIME. Retrieved July 12, 2019, from https://bitesizebio.com/28738/get-genotyping-pcr-work-every-time/
Ap Tech Nova. (2018, April 25). DNA Microarray Technology. Retrieved July 12, 2019, from https://www.youtube.com/watch?v=yzBVOCwRanI
AuLab. (2016, October 22). Retrieved July 12, 2019, from https://www.youtube.com/watch?v=PZW_eyDzYVQ
Bioinformatics, U. (2014, August 13). Bridge Amplification Part 1. Retrieved from https://www.youtube.com/watch?v=3URxJD5k3T8
Blog, R. (2018, April 20). A Workflow Guide to RNA-seq Analysis of Chaperone Function and Beyond. Retrieved July 12, 2019, from https://www.rna-seqblog.com/a-workflow-guide-to-rna-seq-analysis-of-chaperone-function-and-beyond/
Bridge PCR. (n.d.). Retrieved July 12, 2019, from https://binf.snipcademy.com/lessons/ngs-techniques/bridge-pcr
Castel, S. E., Levy-Moonshine, A., Mohammadi, P., Banks, E., & Lappalainen, T. (2015). Tools and best practices for data processing in allelic expression analysis. Genome Biology,16(1). doi:10.1186/s13059-015-0762-6
Dorak, T. (2007, June). Genotyping with PCR. Retrieved July 12, 2019, from https://www.the-scientist.com/lab-tools-old/genotyping-with-pcr-46437
Emulsion PCR ePCR. (n.d.). Retrieved July 12, 2019, from https://binf.snipcademy.com/lessons/ngs-techniques/emulsion-pcr
EncyclopediaBritannica. (2018, December 05). Single nucleotide polymorphism. Retrieved July 12, 2019, from https://www.britannica.com/science/single-nucleotide-polymorphism
FGRS: Protocol RNA-seq Library Preparation. (2011). Retrieved July 12, 2019, from http://fg.cns.utexas.edu/fg/protocol__RNA-seq_Library_Preparation.html
Gene Expression Analysis. (n.d.). Retrieved July, 2019, from https://www.rtsf.natsci.msu.edu/genomics/gene-expression-analysis/
Hadfield, J. (2017, December 07). An Introduction to RNA-seq. Retrieved July 12, 2019, from https://bitesizebio.com/13542/what-everyone-should-know-about-rna-seq/
Hellemans, J. (2019, August 26). Benchmarking RNA sequencing sensitivity using transcriptome-wide RT-qPCR data. Retrieved July 12, 2019, from https://blog.qbaseplus.com/benchmarking-rna-sequencing-sensitivity-using-transcriptome-wide-rt-qpcr-data
Khan, R. (2019). What is genotyping? Retrieved July 12, 2019, from https://help.insito.me/en/articles/859665-what-is-genotyping
Kurdyukov, S., & Bullock, M. (2016, January 06). DNA Methylation Analysis: Choosing the Right Method. Retrieved July 12, 2019, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4810160/
Learn Genetics. (n.d.). DNA Microarray. Retrieved July 12, 2019, from https://learn.genetics.utah.edu/content/labs/microarray/
Little, S. (2001, May 01). Amplification‐Refractory Mutation System (ARMS) Analysis of Point Mutations - Little - 1995 - Current Protocols in Human Genetics - Wiley Online Library. Retrieved July 19, 2019, from https://currentprotocols.onlinelibrary.wiley.com/doi/abs/10.1002/0471142905.hg0908s07
Lutze, K. (2017). 7 Secret Tricks to Optimize Emulsion PCR in the NGS Workflow. Retrieved July 12, 2019, from https://blog.hseag.com/7-secret-tricks-to-optimize-emulsion-pcr-in-the-ngs-workflow
Mackenzie, R. (2018). RNA-seq: Basics, Applications and Protocol. Retrieved July 12, 2019, from https://www.technologynetworks.com/genomics/articles/rna-seq-basics-applications-and-protocol-299461
Morey, J. S., Ryan, J. C., & Van Dolah, F. M. (2006). Microarray validation: Factors influencing correlation between oligonucleotide microarrays and real-time PCR. Retrieved July 12, 2019, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1779618/
NIH. (2019). What are single nucleotide polymorphisms (SNPs)? - Genetics Home Reference - NIH. Retrieved July 12, 2019, from https://ghr.nlm.nih.gov/primer/genomicresearch/snp
NIH. (2015, August 27). DNA Microarray Technology Fact Sheet. Retrieved July, 2019, from https://www.genome.gov/about-genomics/fact-sheets/DNA-Microarray-Technology
Shepherd, C. (2019, June 20). Quantifying Allele-Specific Gene Expression Using PCR-Based Methods. Retrieved July 12, 2019, from https://bitesizebio.com/36768/quantifying-allelle-specific-gene-expression-using-pcr-based-methods/
StatQuest with Josh Starmer. (2017, August 31). Retrieved July 12, 2019, from https://www.youtube.com/watch?v=tlf6wYJrwKY&t=636s
Udacity. (2015, February 23). Retrieved July 12, 2019, from https://www.youtube.com/watch?v=qS8eboruKFE
USD Bioinformatics. (2014, August 15). Emulsion PCR. Retrieved July 12, 2019, from
Wang, Z., Gerstein, M., & Snyder, M. (2009, January). RNA-Seq: A revolutionary tool for transcriptomics. Retrieved July 12, 2019, from https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2949280/
Zou, J. (n.d.). ASE. Retrieved July 12, 2019, from http://genetics.cs.ucla.edu/ase/

The protein streptavidin clamps onto the small molecule biotin in one of the tightest natural interactions ever discovered. Over the years, the strength of this...

Maybe you are about to start working with D-luciferin, and you begin reviewing protocols. But what you’re doing is a little different. Or what you...

Handles are common in everyday life. We use them to help us open doors, cabinets, or to hold coffee mugs, cookware, and tools. They are...

Labeling antibodies with biotin or a fluorophore enables scientists to detect, track, and quantify that antibody and the molecules that it binds to. We cover...
