19 Common Questions About Kanamycin
by Adriana Gallego, Ph.D.

by Adriana Gallego, Ph.D.
This article gives you quick answers to some very common questions when working with kanamycin monosulfate.
Below is a clickable list of the top compiled questions as well as detailed answers to each one.
Take a look and save time in your daily work.
In this article:
1. How does kanamycin inhibit cell growth (Kanamycin mechanism of action)
2. Is kanamycin bacteriostatic or bactericidal?
3. What is the monosulfate in kanamycin monosulfate
4. What is the difference between kanamycin A and kanamycin B?
5. What is the chemical structure of kanamycin?
6. What are the resistance genes for kanamycin?
7. What is the kanamycin (KanR) resistance gene sequence?
8. Why would I want to use the kanamycin resistance gene?
9. Does kanamycin work on gram-negative bacteria or gram-positive bacteria?
10. What kind of antibiotic is kanamycin?
11. What bacteria are kanamycin effective against
12. What concentration of kanamycin should I use for selection?
13. What concentration of kanamycin should I use for transformation?
14. Is kanamycin a good antibiotic for cell culture?
15. What is the difference between kanamycin and other commonly used antibiotics?
16. What are the major uses of kanamycin in biotechnology?
17. How do you add kanamycin to media for selection?
18. How do you make a kanamycin stock solution?
19. How do you make a kanamycin working solution?
Kanamycin inhibits cell growth (mechanism of action) by inhibiting protein synthesis. It irreversibly binds to the 30S ribosome subunit of the bacterium. This causes an incorrect alignment of the mRNA and eventually produces a misread, causing the wrong amino acid to be placed into the peptide chain.

Figure 1. Kanamycin mechanism of action
Consequently, placement of the wrong amino acid into the peptide chain leads to nonfunctional proteins being produced. These nonfunctional, abnormal proteins accumulate in bacteria leading to cell death.
The mechanism of action for kanamycin and any aminoglycoside antibiotic is performed in three steps.
Step 1. Kanamycin enters the cell through a self-promoted uptake mechanism.
Self-promoted uptake involves the displacement of divalent cations from lipopolysaccharides by the polycationic aminoglycosides.
This interferes with cross-bridging between neighboring lipopolysaccharide molecules, which leads to outer membrane destabilization and enhancement of antibiotic uptake.
Afterward, antibiotics like aminoglycosides (e.g. kanamycin) cross the gram-negative outer membrane (OM), they disrupt the inner membrane (IM) which releases internal constituents like Ca2+ and Mg2+.
Therefore, antibiotics can cross both membranes and reach the cytoplasm.
Without magnesium and calcium ions, the outer and inner membranes become more permeable. This in turn, elevates the kanamycin uptake. After crossing both membranes barriers, kanamycin enters the cytoplasm and targets the ribosome.

Figure 2. Kanamycin enters the cell through a self-promoted uptake mechanism in gram-negative bacteria.
The type of membrane components kanamycin interacts with depends on whether bacteria is gram-positive or gram-negative.
Usually, the same ion displacement occurs in both gram-positive or gram-negative to facilitate the antibiotic entrance.
For gram-positive bacteria, kanamycin interacts with phospholipids and teichoic acids.
For gram-negative bacteria, kanamycin binds to the phospholipids and lipopolysaccharides in the cell membrane.
Step 2. In the second step, kanamycin binds to the A-site on the 16S RNA of the 30S bacterial ribosome hindering the normal translation process, which leads to mistranslated proteins in the cytoplasm.
These mistranslated proteins accumulate and damage the cytoplasmic membrane.
As damage occurs to the cytoplasmic membrane, more kanamycin enters, which increases protein synthesis inhibition and accelerates cell death (Kause et al 2016).

Kanamycin is a bactericidal antibiotic, meaning it kills bacteria. Kanamycin binds to the 30S subunit of the bacterial ribosome, which interferes with the translation process, causing misreads during protein synthesis. Abnormal proteins accumulate, causing bacterial cell death.
Bacteriostatic antibiotics, on the other hand, are commonly described as substances that arrest growth but do not kill the pathogen.
However, there is a deeper difference between bacteriostatic and bactericidal antibiotics that depends on their concentrations and the effect over bacteria.
Before explaining that difference I should present two terms.
MIC indicates the lowest level of antimicrobial agent that greatly inhibits growth, and it is physically visualized as a tube with very low turbidity.
If, for example you were to plate an aliquot from this tube you will be able to visualize some bacterial growth.
MBC, on the other hand, indicates the lowest level of antimicrobial agent resulting in microbial death and it is physically visualized as a tube with no turbidity.
If you were to plate an aliquot from this tube you won’t see bacterial growth.

Figure 4. Illustrates the difference between MBC and MIC.
A formal difference between bactericidal and bacteriostatic is bactericidal antibiotics have a ratio of MBC to MIC ≤ 4, while a bacteriostatic antibiotic has an MBC to MIC ratio of > 4.
From our example, the MBC is 8 µg/ml and the MIC is 4 µg/ml, so the ratio of MBC to MIC is:
(MBC)/ (MIC)= (8 µg/ml) / (4 µg/ml) = 2As MBC to MIC ≤ 4, the antibiotic is bactericidal.
In this sense, bacteriostatic antibiotics also kill bacteria; they just require a higher concentration than bactericidal agents to achieve specific thresholds of bacterial reduction.
Examples of bacteriostatic antibiotics include chloramphenicol, clindamycin, and linezolid.
Published, randomized and controlled trials demonstrated bactericidal agents are not intrinsically superior in efficacy to bacteriostatic agents.
A study reported by (Dickler et al 2017) analyzed 56 trials that compared bacteriostatic with bactericidal, and authors showed that the majority of trials across a variety of infections found no difference in efficacy between bacteriostatic versus bactericidal agents.
Antibiotics are often chemically made into their salt forms to enhance their solubility, absorption, and effectiveness.
Kanamycin monosulfate is the sulfate salt form of kanamycin (Figure 4). Monosulfate indicates there is only one salt type in the reagent. There are other reagents where two salts (disulfate/bisulfate) are present. The salt forms of the antibiotic enhance solubility, absorption, and effectiveness.
Sulfate (SO₄²-) is an inorganic ion that binds to the molecules favoring their chemical properties such as solubility, absorption, and effectiveness.
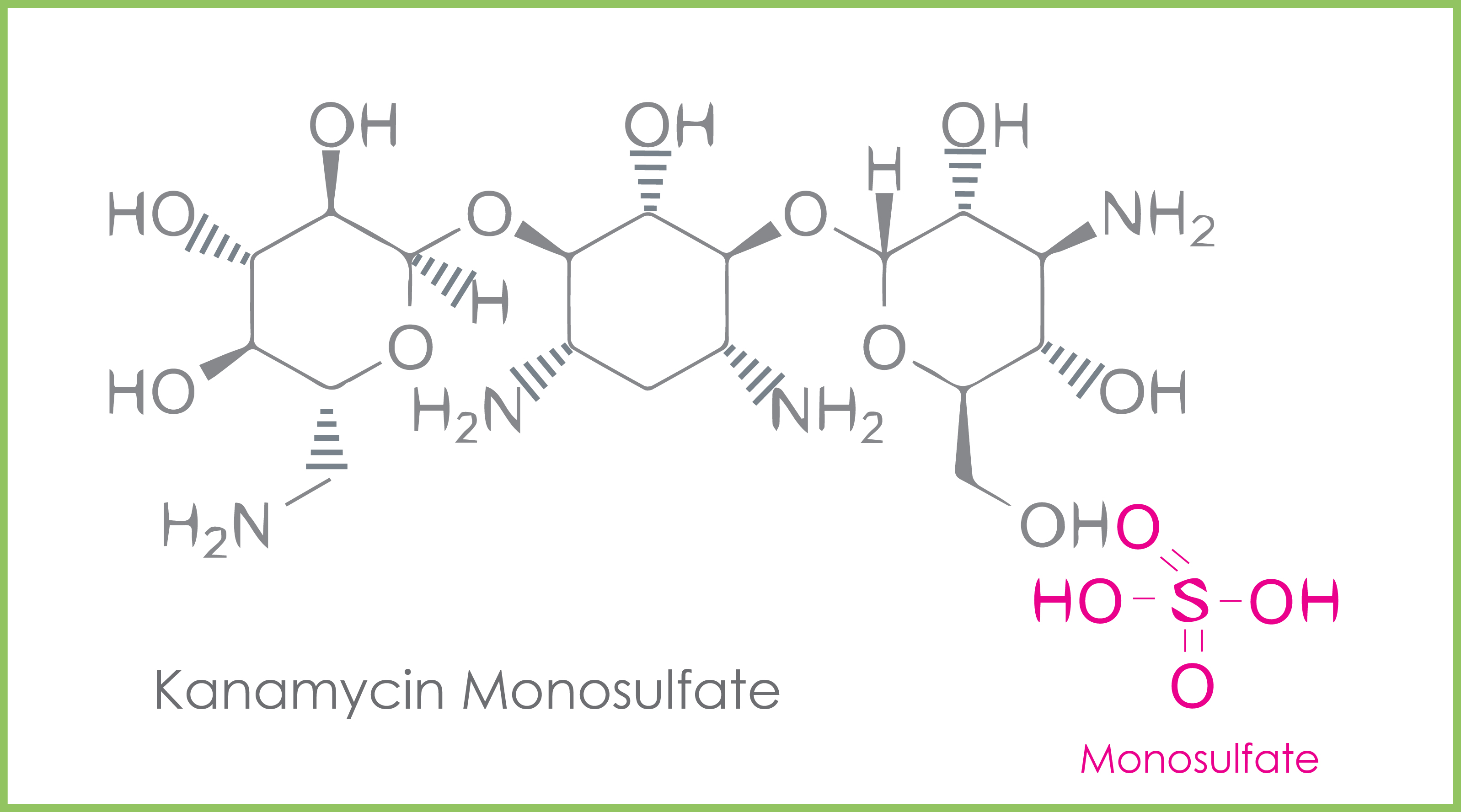
Figure 5. The molecular structure of kanamycin monosulfate.
The difference between kanamycin A and kanamycin B is the chemical group in the C2’ position. There is a hydroxy group at position C2 in kanamycin A; whereas, kanamycin B has an amino group at the C2 position.

Figure 6. Highlights the two different chemical groups in the C2’ position of kanamycin B and kanamycin A. Kanamycin A is a product of kanamycin B.
Kanamycin A is the main product of kanamycin B. The synthesis of kanamycin A from kanamycin B is catalyzed by KanJ and KanK in Streptomyces kanamyceticus.
Furthermore, due to differences in their C2′ substituents, kanamycin B is less biologically active and has stronger side effects than kanamycin A.
Consequently, kanamycin A is used more often (Gao et al 2017).
Kanamycin has a core structure consisting of amino sugars connected to a dibasic aminocyclitol (commonly 2-deoxystreptamine) via glycosidic linkages. Kanamycin is a 4,6-di-substituted deoxystreptamine ring.

Figure 7. Chemical structure of kanamycin.
The kanamycin resistance gene is NPTII. This gene is abbreviated as KanR in the plasmids. Enterococci and Staphylococci produce an enzyme called aminoglycoside 3'-phosphotransferase also called kanamycin kinase or neomycin-kanamycin phosphotransferasefrom the NPTII gene.
This enzyme inactivates kanamycin by catalyzing the transfer of the terminal phosphate of ATP to kanamycin, a process called phosphorylation.

Figure 8. NPTII catalyzes the transfer of the terminal phosphate on ATP (Pink) to kanamycin via phosphorylation. This enzymatic action inactivates kanamycin.
The NPTII gene is introduced in vectors which are used in genetic engineering to select correctly transformed bacterial organisms.
Those bacteria growing in a medium with kanamycin can do so because they harbor the NPTII gene that inactivates kanamycin.
The kanamycin resistance gene is found in the commercial pUC vectors pHSG298 and pHSG299.
The resistance gene (KanR) is 816bp, 31kDa and coded for an aminoglycoside 3'-phosphotransferase also called kanamycin kinase.

Figure 9. Vector map showing the KanR gene available in commercial plasmids of pHSG29 and pHSG299.
There are many kanamycin resistance gene sequences available in the UniProt database (5215 results). We provide some sequences for E. coli, Bacillus cereus, and Pseudomonas fluorescens. Depending on the plasmid, you may have a different sequence for KanR.
While there are many kanamycin resistance gene sequences, we provide one example of a gene sequence for E. coli.
SEQUENCE256 AA;29166 MW;B45EF8CDBD00654E CRC64;
MRIVNGPIIM TREERMKIVH EIKERILDKY GDDVKAIGVY GSLGRQTDGP YSDIEMMCVM
STEEAEFSHE WTTGEWKVEV NFDSEEILLD YASQVESDWP LTHGQFFSIL PIYDSGGYLE
KVYQTAKSVE AQTFHDAICA LIVEELFEYA GKWRNIRVQG PTTFLPSLTV QVAMAGAMLI
GLHHRICYTT SASVLTEAVK QSDLPSGYDH LCQFVMSGQL SDSEKLLESL ENFWNGIQEW
TERHGYIVDV SKRIPF
Below is a kanamycin resistance gene sequence for Bacillus cereus, though many sequences exist.
SEQUENCE293 AA;33883 MW;DF9E319CD3E5041C CRC64;
MRKENEYVTY LQRLYPDLQI NSVYINEIGQ NNDVLIVNDN IVFRFPKYEK GIQKLRIETQ
LLEKIRPFIT LQIPNPSYQG FQNEVPGKVF AGYEMIEGDP FWKNVFTEIN DEKRLQKLAC
TLARFLKELH EIPLSTFEGI MQCDSTDMYS EINSLYSQLK EHVYPFMRNV ARKEVSTSFE
LYLNESSHFN FTPSLVHGDF GMTNILYSAT KKNISGVIDF GGASIGDPAY DFAGILASYG
EEFLQLFEAY YPNLEAVKER MYFYKSTFAL QEALFGVLNN DKKAFEAGIA QYV
MEARLVDQEE LDEEHRGLSP ALLFERLLNR QPAEQDLVVT HGDACMPNII VDDGRFTGFI
DCGRLGVADR YQDLALATRD IAEELGEQWI APFLRRYAVL EADQDRIYFY RLLDEFF
Tip: To help you find the proper KanR of your plasmid, click in this link from Uniprot database, you’ll land on the “kanamycin kinase” term. From there, you may select your organism of interest.
Kanamycin is good for selection because it is a bactericidal antibiotic.
Antibiotics with bactericidal functions substantially kill a pathogen compared to bacteriostatic antibiotics which arrest the bacterial growth. Other reasons to use the kanamycin resistance gene or any other depend on the resistance gene already present in the plasmid backbone. If the vector comes by default with KanR, you should use kanamycin as a selection antibiotic.
Kanamycin is mainly effective against gram-negative bacteria. But it is also effective against some gram-positive bacteria.
Kanamycin is an aminoglycoside antibiotic. The role of aminoglycosides is to stop bacteria from producing proteins needed for their survival. Aminoglycosides are useful primarily in infections involving aerobic, and gram-negative bacteria.
Because aminoglycosides inhibit protein production, bacterial cells die, which therefore classifies the aminoglycoside family of antibiotics as bactericidal.
The core structure of aminoglycosides is composed of amino sugars connected to a dibasic aminocyclitol (commonly 2-deoxystreptamine) via glycosidic linkages.
Scientists have categorized aminoglycosides into four classes based on the nature of the aminocyclitol moiety:
Ref: Krause et al 2016.

Figure 11. Types of antibiotics
Kanamycin is effective against gram-positive bacteria like:
Kanamycin is effective against gram-negative bacteria such as:
Generally, the concentration of kanamycin used for selection with plasmids is 50 µg/ml, and 20 µg/ml for cosmids. However, it is always advisable to test different concentrations starting from 10 µg/mL, 20 µg/mL, 50 µg/mL, 75 µg/mL and 100 µg/mL to evaluate if cells still survive or die.
The recommended concentration of kanamycin for transformation is 50 µg/ml. However, it is highly advisable to test different concentrations starting from 10 µg/mL to 100 µg/mL to evaluate if cells still survive or die.
Kanamycin is a good antibiotic for cell culture because it is bactericidal, which causes cell lysis. Kanamycin is superior to bacteriostatic antibiotics because bacteriostatic antibiotics only arrest bacterial growth but do not kill them.
Furthermore, the activity of kanamycin, and aminoglycosides in general, can be enhanced in vitro through synergy with other antibiotics like beta-lactams (e.g., carbenicillin, penicillin G, ampicillin) against gram-positive and gram-negative microorganisms (Kause et al 2016).
Kanamycin differs from other commonly used antibiotics in the structure, the antibiotic effect (bacteriostatic or bactericidal), or the mechanism of action.
There are two key differences between kanamycin and ampicillin: antibiotic class and their mechanisms of action. Kanamycin is an aminoglycoside whose mechanism of action is to inhibit protein synthesis. Ampicillin is a beta-lactam whose mechanism is inhibiting cell wall synthesis.
The difference between kanamycin and gentamycin is their antibacterial effect. According to Kwang (1985), kanamycin is less bactericidal in vivo and lyses less bacteria (bacteriolysis) and endotoxins compared to gentamycin, which reduces early death incidence.
The difference between kanamycin and neomycin is their chemical structure. While kanamycin has a 4,6-di-substituted deoxystreptamine ring, neomycin has a 4,5-di-substituted deoxystreptamine ring. In general, neomycin is used in experiments on prokaryotic cells while kanamycin can be used also for eukaryotic cells.
Kanamycin and neomycin are structurally different but mechanistically similar because they belong to the same antibiotic family, aminoglycosides.
Kanamycin can be used in biotechnology to prevent bacterial contamination in cell cultures. In molecular biology, kanamycin is used as a selectable marker to select colonies successfully transformed with the kanamycin resistance gene.
It can be also used to select genetically modified plants via agrobacterium transformation harboring the kanamycin resistance gene.
Use a pipette to add 1 ml of kanamycin stock solution (50 ml/L) to 1 L of culture media to end with a final concentration of 50 µg/ml. Pour about 25 mL into agar plates and then seal the plates to avoid evaporation.
To prepare a kanamycin stock solution of 50 mg/L, first, weigh 500 mg of Kanamycin monosulfate and mix with 10 ml of sterile water. Mix thoroughly to dissolve with the help of a magnetic stir bar. Use a 0.2 µm prewet filter to filter the stock solution. Make 1 ml aliquots and store them at 2-8°C for long term storage.
How to prepare a kanamycin stock solution 50 mg/ml
To know how to test an antibiotic effect using a disk diffusion assay please check our GoldBio video.
Tip: Another quick option is to use ready-made solutions like our kanamycin stock solution 50mg/mL
To prepare a kanamycin working solution of 50 µg/ml, take 100 ul of a 50 mg/L stock solution and mix with 100 ml of culture media.
How to prepare a kanamycin working solution 50 µg/ml
Gao, W., Wu, Z., Sun, J., Ni, X., Xia, H., 2017. Modulation of kanamycin B and kanamycin A biosynthesis in Streptomyces kanamyceticus via metabolic engineering. PLoS One 12, 1–19. https://doi.org/10.1371/journal.pone.0181971
Gray, G.S., Fitch, W.M., 1983. Evolution of antibiotic resistance genes: The DNA sequence of a kanamycin resistance gene from Staphylococcus aureus. Mol. Biol. Evol. 1, 57–66. https://doi.org/10.1093/oxfordjournals.molbev.a040...
Kim, K.S., 1985. Comparison of gentamicin and kanamycin alone and in combination with ampicillin in experimental escherichia coli bacteremia and meningitis. Pediatr. Res. 19, 1152–1155. https://doi.org/10.1203/00006450-198511000-00007
Ninfa, A.J., Selinsky, S., Perry, N., Atkins, S., Xiu Song, Q., Mayo, A., Arps, D., Woolf, P., Atkinson, M.R., 2007. Using Two-Component Systems and Other Bacterial Regulatory Factors for the Fabrication of Synthetic Genetic Devices. Methods Enzymol. 422, 488–512. https://doi.org/10.1016/S0076-6879(06)22025-1
Recht, M.I., Puglisi, J.D., 2001. Aminoglycoside resistance with homogeneous and heterogeneous populations of antibiotic-resistant ribosomes. Antimicrob. Agents Chemother. 45, 2414–2419. https://doi.org/10.1128/AAC.45.9.2414-2419.2001

Handles are common in everyday life. We use them to help us open doors, cabinets, or to hold coffee mugs, cookware, and tools. They are...

Labeling antibodies with biotin or a fluorophore enables scientists to detect, track, and quantify that antibody and the molecules that it binds to. We cover...

Covalently conjugating a small molecule to an antibody’s surface is a process called antibody “labeling.” Labeling antibodies with small molecules such as biotin or fluorophores...

DNA gel electrophoresis is one of the most widely used analytical techniques in molecular biology, providing a simple and reliable way to separate DNA fragments...
