Beyond pH 7 - Biologically Relevant pH In Diverse Environments
by Simon Currie, Ph.D.

by Simon Currie, Ph.D.
In Flatland, a novel by Edwin Abbott, a square narrates his flat two-dimensional world and his dreams of visiting the more limited, one-dimensional Lineland. In brief, Flatland is a social satire and a critique of the artificial limitations that Victorian society imparted on its inhabitants. Meaning there were many dimensions of people’s lives, especially women, that they were restricted from accessing for fear of being seen as improper.
Why am I bringing up a social critique in an article about biological pH? Because there is one dimension in particular that is dreadfully under explored in biological experiments: pH! Most experimenters rigidly examine their systems at neutral pH. In contrast, the natural world, in which proteins evolved, samples a much richer pH profile (Casey et al, 2010; Decker et al, 2021; Gaohua et al, 2021; Rampelotto, 2013).
Biologically relevant pH is frequently stated to be 7.0 to 7.4, which corresponds to mammalian cytoplasm in healthy growing cells. However, extremophile microbes, organelles, human cells and fluids, stressed cells, and diseased cells exist in diverse environments ranging from at least pH 2 to 13.
Now as a disclosure, for most of my scientific career I also rigidly stuck to neutral pH conditions. When I purified proteins in the lab, I tried to keep my conditions close to pH 7 or 8, and I worried that if I veered too far off into the pH wilderness that my protein would disintegrate into a hundred little pieces.
However, over time I’ve learned that the pH in my experiments shouldn’t follow a standard convention, but rather should be custom fit to the natural needs of my protein of interest. Follow along with me in this article as we explore the interesting biology of proteins finding their niche in a broad range of environments outside of the standard pH 7.
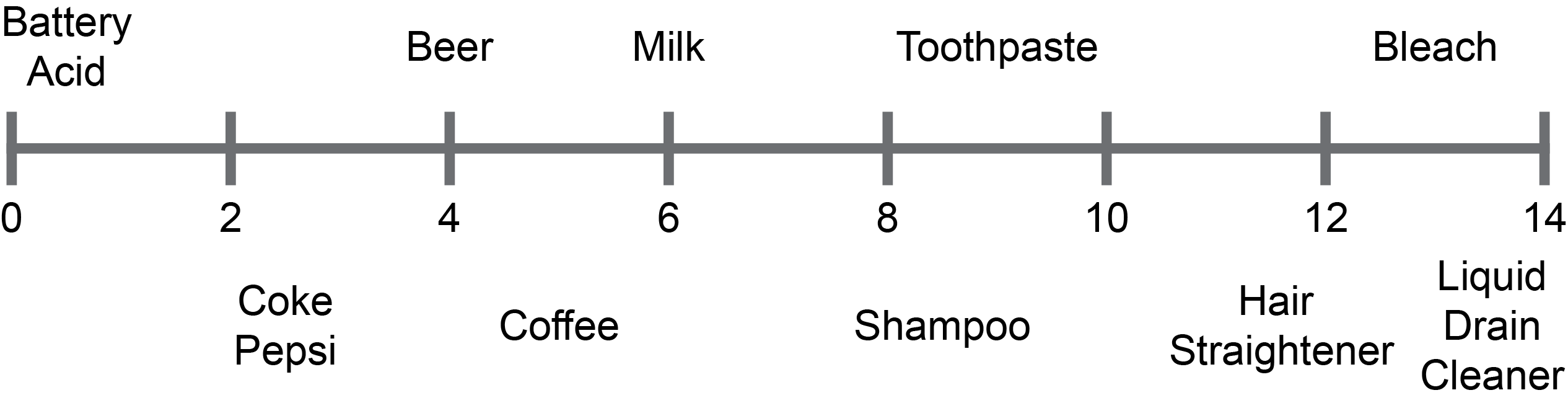
Figure 1. The pH range of common items.

Figure 2. Lake Albert is an alkaline lake in Lake County,
Oregon.
Microorganisms like bacteria can live in drastically different pH environments. For example, some bacteria live in super acidic ocean vents (pH ~ 3) whereas other bacteria thrive in alkaline lakes (pH ~ 13) such as Lake Albert in the desolate Oregon high desert (Rampelotto, 2013).
Hydrogen ion (H+) pumps, which we’ll discuss in the Organelles section, neutralize the intracellular pH of these bugs a little bit, but their internal pH still ranges from approximately 4.5 to 8.5. Remember, as a log scale that 4-pH unit range represents a 10,000 (104) fold difference in H+ ion concentration.
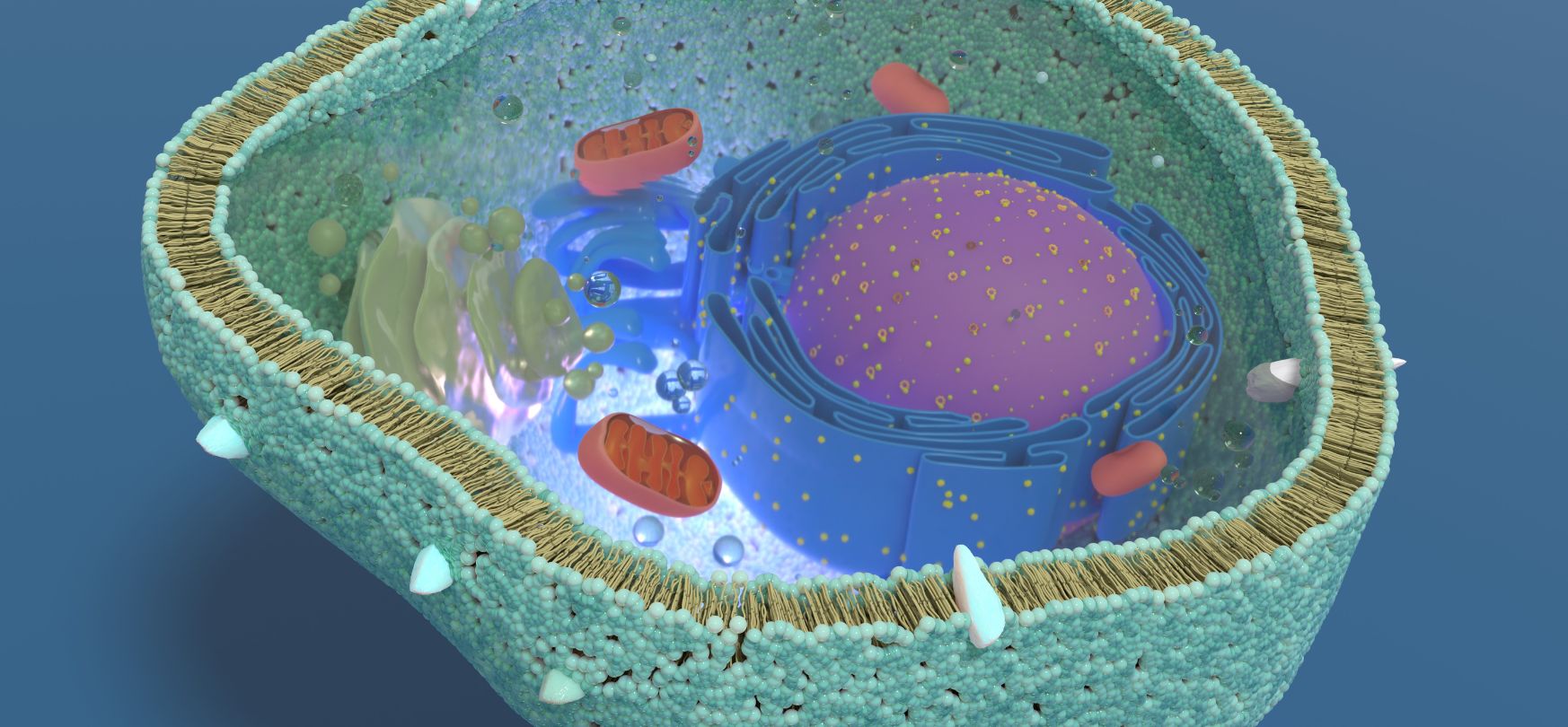
Figure 3. Shows a cell’s cross-section exposing the
organelles. Organelles can have natural pH ranges that differ from a neutral pH
of ~7 (Table 1).
Cells and some organelles have hydrogen ion (H+) pumps that control their internal pH. As a reminder, organelles are membrane-bound structures found within eukaryotic cells. The nucleus, mitochondria, and lysosomes are examples of organelles.
In the absence of hydrogen ion pumps, the pH would equalize across membranes. That is because H+ ions, like any chemical matter, flow from areas of high concentration to low concentration until an equilibrium is reached. We call the discrepancy of molecular concentrations between two given areas a chemical gradient (Figure 4).
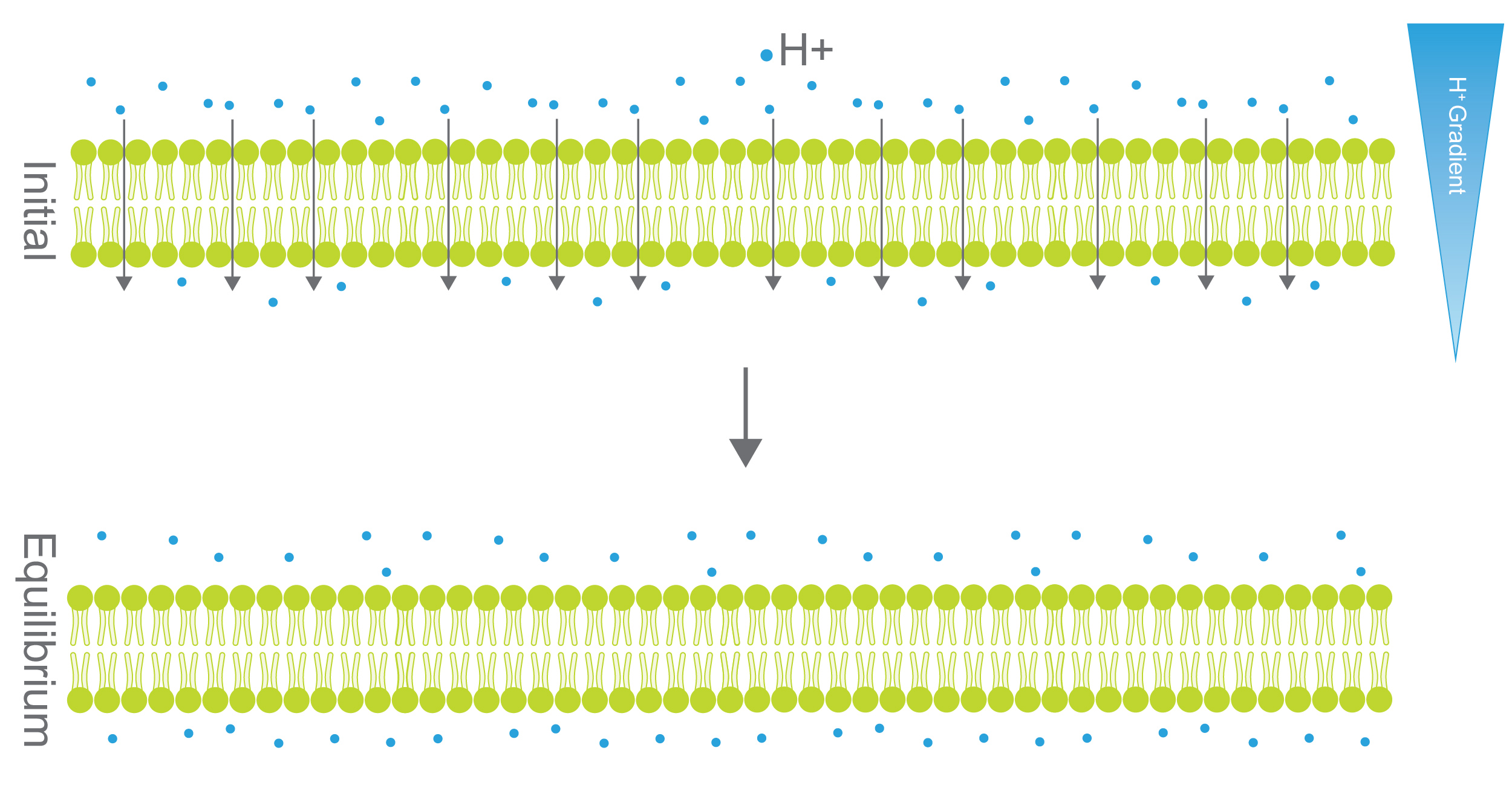
Figure 4. In the top panel, there is a hydrogen ion (H+) gradient across the membrane. Since H+ is small enough to travel through the membrane it will, on average, move from the higher concentration side to the lower concentration side until an equilibrium is reached (right). H+ pumps (Figure 5) prevent this equilibrium process to maintain a H+ gradient.
Yet, when these proton pumps are turned on, they use energy to pump H+ ions against their chemical gradient (Figure 5).
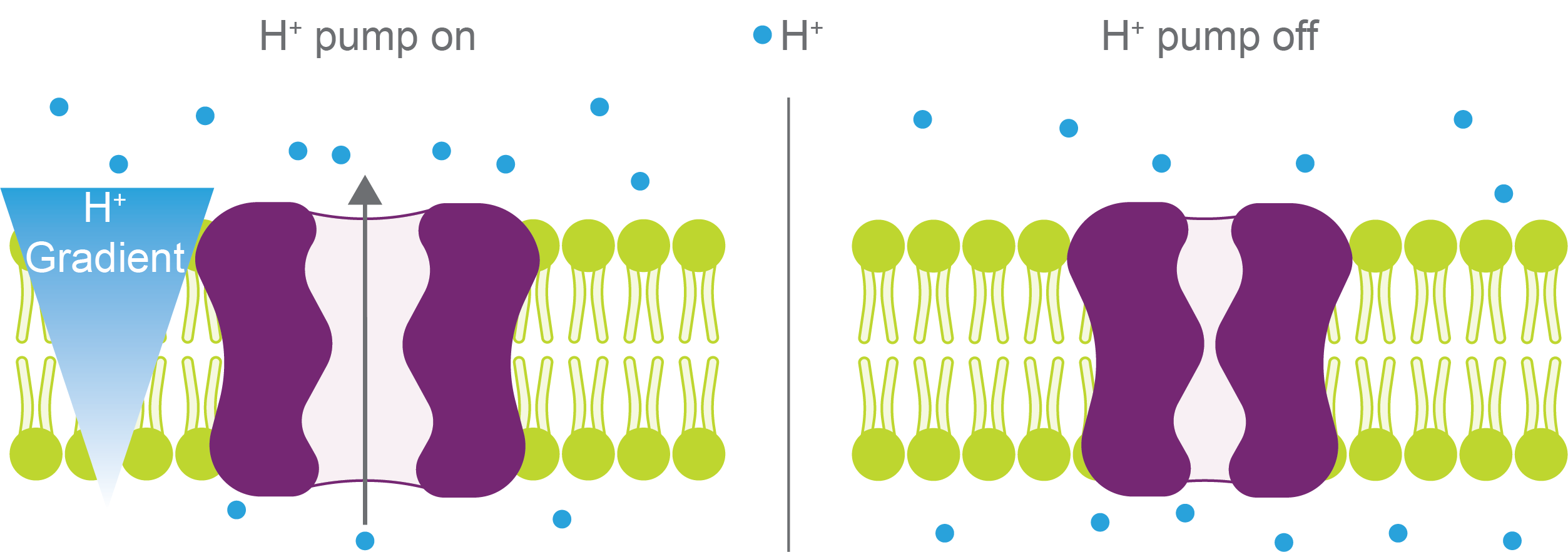
Figure 5.
Hydrogen ion pumps (purple) maintain a hydrogen (blue) gradient across a
membrane (lime green) when turned on (left). When H
+ pumps are off
(right), hydrogen will equilibrate across the membrane.
However, those pumps can rapidly turn off in response to environmental stresses. When this happens, the pH can change several units in a matter of seconds to a few minutes (Joyner et al, 2016; Munder et al, 2016).
These proton pumps maintain the cytoplasm and organelles at different pHs (Table 1). Mitochondria, for example, are relatively basic (pH = 8.0), whereas lysosomes are acidic (pH = 4.7) (Casey et al, 2010; Theillet et al, 2014).
Lysosomes are the recycling center of cells. They break down biopolymers such as proteins, nucleic acids, carbohydrates, and lipids into smaller building blocks that are then used to rebuild new biopolymers. Its acidic pH activates degrading enzymes within the lysosome allowing them to efficiently perform their molecular recycling functions (Bond & Butler, 1987).
Table 1. pH of organelles.
|
Organelle |
pH |
|
Cytoplasm |
7.2 – 7.4 |
|
Nucleus |
7.2 – 7.4 |
|
Mitochondria |
8.0 |
|
Endoplasmic Reticulum |
7.5 |
|
Golgi Apparatus |
6.6 |
|
cis-Golgi Cisterns |
6.7 |
|
trans-Golgi Networks |
6.0 |
|
Peroxisomes |
7.0 |
|
Secretory Granules |
5.5 |
|
Early Endosomes |
6.3 |
|
Recycling Endosomes |
6.5 |
|
Late Endosomes |
5.5 |
|
Lysosomes |
4.7 |
Values from Casey et al, 2010 and Theillet et al, 2014
So far in this article, we’ve been thinking at the level of single cells. In the cases of extremophile bacteria and eukaryotes like brewer’s yeast, the single cell is the whole organism. However, in multicellular organisms, many cells come together to make the entire organism. Humans, for example, have trillions of cells with many different specialized cell types like stomach, intestine, and heart cells (Hatton et al, 2023) (Figure 6).
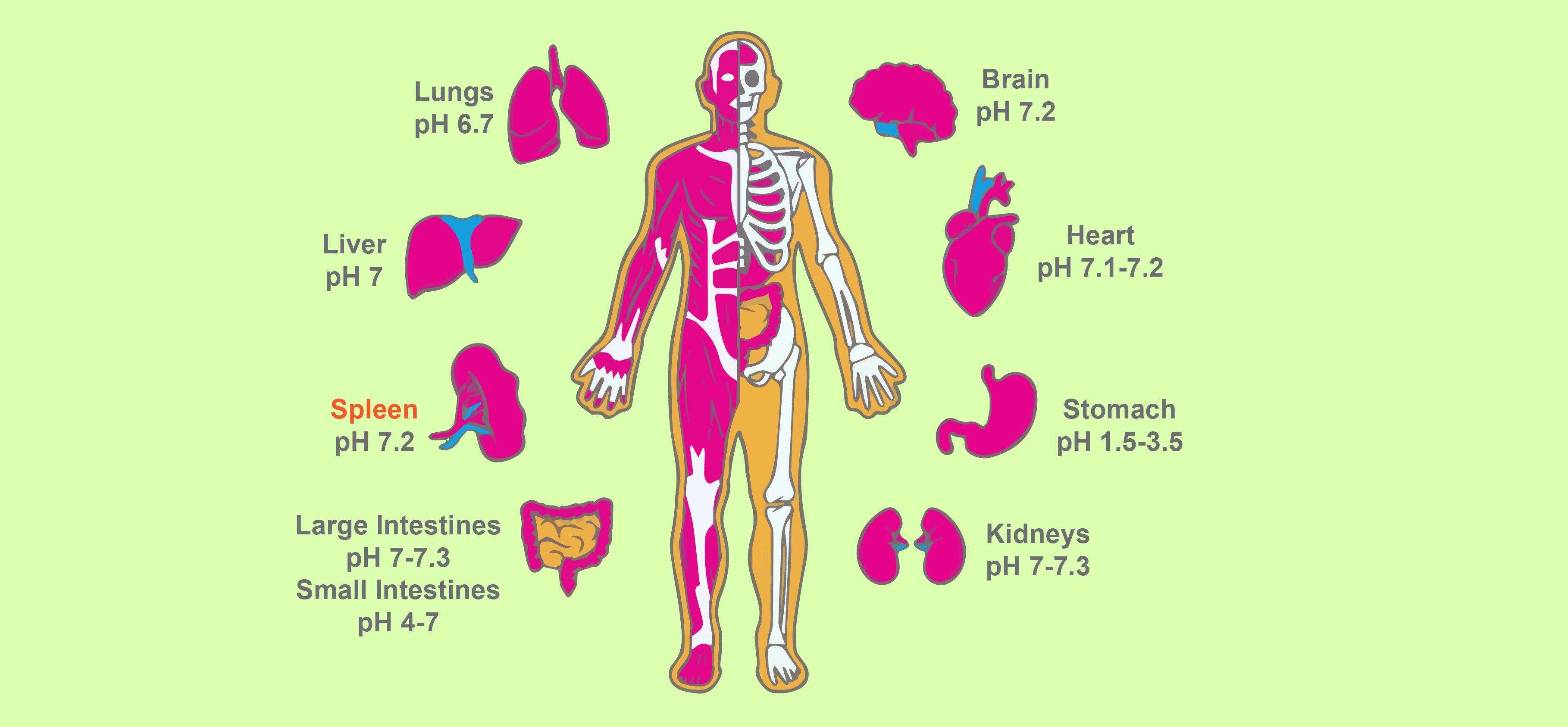
Figure 6. Diagram of major human organs with the general pH of each organ labeled.
You probably already know that the inside of our stomach is very acidic (pH = 2). In contrast, our pancreas is slightly basic (pH = 8). These are more of the edge cases in terms of human cells, but different cells have a variety of pH values within this range (Gaohua et al, 2021) (Table 2).
Table 2. pH of human organs and fluids.
|
Organ or Fluid |
pH |
|
Lungs |
6.7 |
|
Brain |
7.2 |
|
Heart |
7.1 – 7.2 |
|
Liver |
7.0 |
|
Stomach |
1.5 – 3.5 |
|
Gastric Fluid |
2 – 3 |
|
Small Intestine |
4 – 7 |
|
Large Intestine |
5.5 – 7.0 |
|
Spleen |
7.2 |
|
Kidneys |
7.0 – 7.3 |
|
Testis |
7.3 |
|
Semen |
7.2 – 8.0 |
|
Uterus |
6.6 – 7.6 |
|
Vaginal secretions |
3.5 – 4.0 |
|
Bone |
6.3 |
|
Saliva |
6.5 |
|
Pancreas |
5.5 |
|
Pancreatic Fluid |
7.5 – 8.8 |
|
Bladder |
4.7 |
|
Urine |
4.8 – 7.5 |
|
Sweat |
4.0 – 6.8 |
|
Tears |
6.5 – 7.6 |
Values from Gaohua et al, 2021
These pH values are important in organ function, and enzymes have evolved to be active at the pH of the organ where they reside or are active. In the stomach, for example, the very acidic environment helps to break down the proteins and other biopolymers that we eat. Acidic pH also activates pepsin, the primary stomach digestive enzyme that cuts up dietary proteins (Figure 7). In fact, in a test tube, pepsin is most active at pH 2, which perfectly matches the pH of the stomach!
But why is it stable and active at pH 2? Pepsin is chock full of the acidic residues aspartate and glutamate which make up nearly 15% of the protein (Campos & Sancho, 2003). This may be why, contrary to our neutral pH-centric intuitions, pepsin is very stable at pH 2, but is irreversibly denatured (meaning it doesn’t function as a protease anymore) at neutral pH (7 and above).
Two particular aspartate residues are at the active site of pepsin and are very important for its activity: Asp32 and Asp215 (Cornish-Bowden & Knowles, 1969; Fujinaga et al, 1995; Lin et al, 1992) (Figure 7). At least one of these residues needs to be protonated for pepsin to be active. If you read our article on how to calculate protein charge, you’ll learn what a pKa is and that the pKa of aspartate residues is roughly 4. You’ll also learn that for aspartate to be protonated the pH needs to be a bit (1-2 pH units) more acidic than 4. That’s the molecular basis for pepsin having optimal protease activity at pH 2.
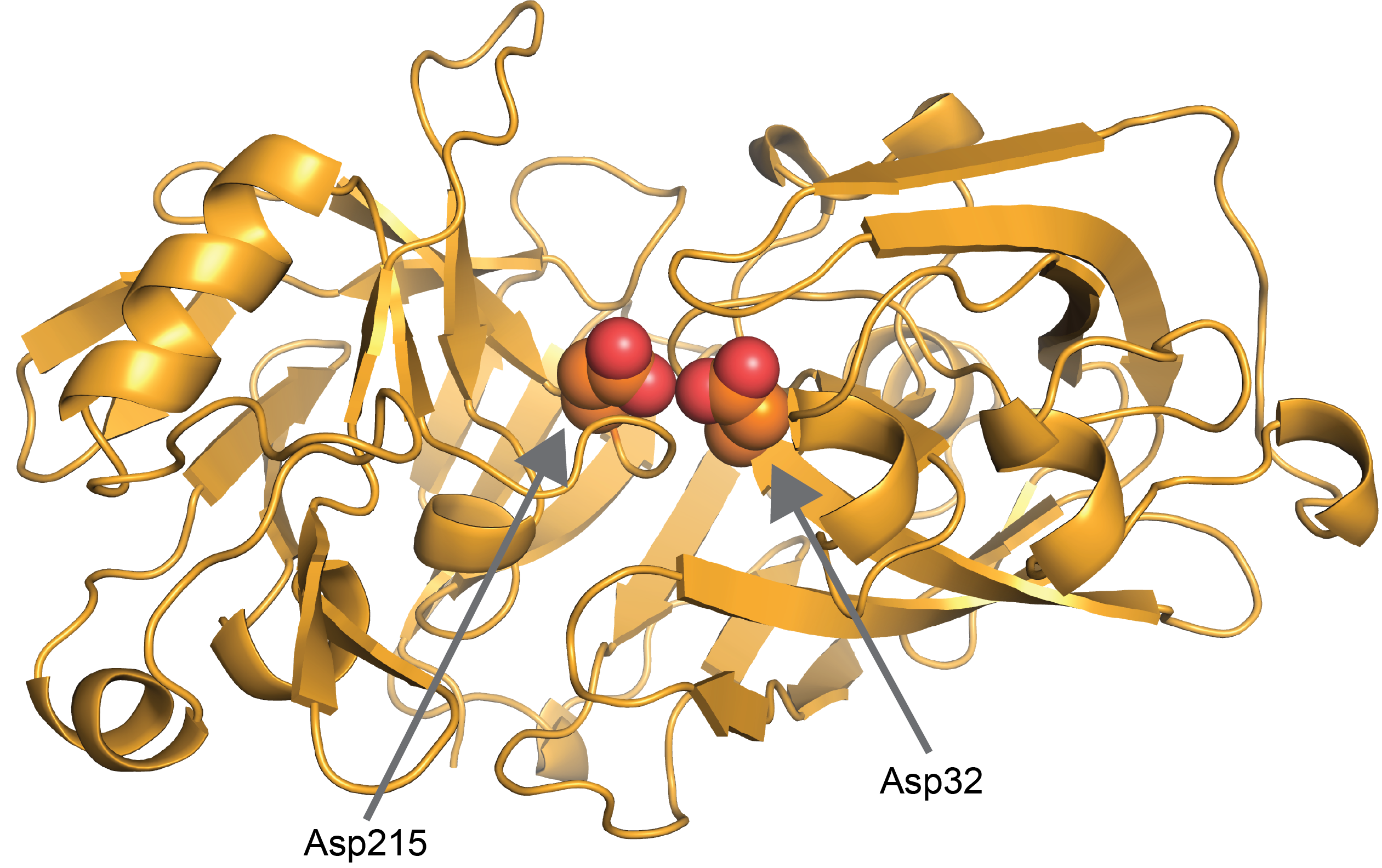
Figure 7. Two catalytic aspartic acids, Asp32 and
Asp215, are at the center of pepsin’s active site. At least one aspartic acid
has to be protonated which is why pepsin is active at very acidic pH in the
stomach (PDB: 1PSN).
Once the digested food and pepsin travel to the duodenum, the pH changes to 6 and pepsin is inactivated. But there are other enzymes (trypsin, chymotrypsin, elastase, and carboxypeptidase) in the duodenum that have optimized activity in this pH and take over the digestion process for pepsin (Heda et al, 2023).
As you can see, this is a really cool example of how having different pH environments in different organs regulates molecular processes to have the right enzyme turned on in the appropriate location. And you can also see how using a neutral pH of 7 would never yield active pepsin.
How do changes in pH lead to differences in biological functions? Proteins “sense” the change in pH by a change in protonation state on one or more amino acids which impacts the protein’s activity, interactions, or stability. As we discussed above in the case of pepsin those sensors are Asp32 and Asp 215 (Figure 7).
By the way, if you need a quick refresher on ionizable amino acids and how pH impacts their charge we have just the article for you.
Proteins that change in response to the surrounding pH are often called pH switches because the pH switches them from one state to another. For example, the acidic environment of the stomach switches pepsin from an inactive state to an active one. Changing enzyme activity is only one way that pH switches work. In addition to this, pH mediates changes in proteins by:
So far, we’ve covered pH changes in different spatial locations such as distinct organs, organelles, and even habitats such as Lake Albert. But pH can also change across timescales.
For example, the cytoplasm becomes slightly more acidic as humans age (pH 6.8 – 7.1) (Decker et al, 2021). This geriatric cytoplasmic acidification is conserved all the way from brewer’s yeast (Saccharomyces cerevisiae) to humans (Homo sapiens)(Choi et al, 2021), and may contribute to cell death and the aggregation of proteins in human neurodegenerative diseases.
pH also changes due to diseases such as cancer. Relative to normal cells, the intracellular pH of cancer cells is slightly basic (7.3 – 7.6) whereas the extracellular pH is more acidic (6.8 – 7.0) (White et al, 2017). Changes enacted by acidic pH that are thought to enable cancer progression include:
These changes are mediated by pH switches similar to those described above.For more details on these protein switches and how slight pH differences drive oncogene progression, take a look at this article.
As you’ve seen in this article, biologically relevant pH covers most of the pH range. So next time someone tells you that biological pH is 7.2, you will be well prepared with your “well, actually …” response.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The Protein Data Bank. Nucleic acids research, 28(1), 235–242. https://doi.org/10.1093/nar/28.1.235
Berman, H., Henrick, K., & Nakamura, H. (2003). Announcing the worldwide Protein Data Bank. Nature structural biology, 10(12), 980. https://doi.org/10.1038/nsb1203-980
Bond JS, Butler PE. Intracellular proteases. Annu Rev Biochem. 1987;56:333-64. doi: 10.1146/annurev.bi.56.070187.002001. PMID: 3304137.
Campos, L. A., & Sancho, J. (2003). The active site of pepsin is formed in the intermediate conformation dominant at mildly acidic pH. FEBS letters, 538(1-3), 89–95. https://doi.org/10.1016/s0014-5793(03)00152-2
Casey, J. R., Grinstein, S., & Orlowski, J. (2010). Sensors and regulators of intracellular pH. Nature reviews. Molecular cell biology, 11(1), 50–61. https://doi.org/10.1038/nrm2820
Chiariello, M. G., Grünewald, F., Zarmiento-Garcia, R., & Marrink, S. J. (2023). pH-Dependent Conformational Switch Impacts Stability of the PsbS Dimer. The journal of physical chemistry letters, 14(4), 905–911. https://doi.org/10.1021/acs.jpclett.2c03760
Choi J., Wang, S., Li, Y., Hao, N., Zid, B. M. (2021). Age-induced P-bodies become detrimental and shorten the lifespan of yeast. bioRxiv 2021.11.05.467477; doi: https://doi.org/10.1101/2021.11.05.467477
Cornish-Bowden, A. J., & Knowles, J. R. (1969). The pH-dependence of pepsin-catalysed reactions. The Biochemical journal, 113(2), 353–362. https://doi.org/10.1042/bj1130353
Currie, S. L., Xing, W., Muhlrad, D., Decker, C. J., Parker, R., & Rosen, M. K. (2023). Quantitative reconstitution of yeast RNA processing bodies. Proceedings of the National Academy of Sciences of the United States of America, 120(14), e2214064120. https://doi.org/10.1073/pnas.2214064120
Czowski, B. J., Romero-Moreno, R., Trull, K. J., & White, K. A. (2020). Cancer and pH Dynamics: Transcriptional Regulation, Proteostasis, and the Need for New Molecular Tools. Cancers, 12(10), 2760. https://doi.org/10.3390/cancers12102760
Decker, Y., Németh, E., Schomburg, R., Chemla, A., Fülöp, L., Menger, M. D., Liu, Y., & Fassbender, K. (2021). Decreased pH in the aging brain and Alzheimer's disease. Neurobiology of aging, 101, 40–49. https://doi.org/10.1016/j.neurobiolaging.2020.12.0...
Fujinaga, M., Chernaia, M. M., Tarasova, N. I., Mosimann, S. C., & James, M. N. (1995). Crystal structure of human pepsin and its complex with pepstatin. Protein science : a publication of the Protein Society, 4(5), 960–972. https://doi.org/10.1002/pro.5560040516
Gaohua, L., Miao, X., & Dou, L. (2021). Crosstalk of physiological pH and chemical pKa under the umbrella of physiologically based pharmacokinetic modeling of drug absorption, distribution, metabolism, excretion, and toxicity. Expert opinion on drug metabolism & toxicology, 17(9), 1103–1124. https://doi.org/10.1080/17425255.2021.1951223
Hatton, I. A., Galbraith, E. D., Merleau, N. S. C., Miettinen, T. P., Smith, B. M., & Shander, J. A. (2023). The human cell count and size distribution. Proceedings of the National Academy of Sciences of the United States of America, 120(39), e2303077120. https://doi.org/10.1073/pnas.2303077120
Heda R, Toro F, Tombazzi CR. Physiology, Pepsin. [Updated 2023 May 1]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK537005/
Lin, Y., Fusek, M., Lin, X., Hartsuck, J. A., Kezdy, F. J., & Tang, J. (1992). pH dependence of kinetic parameters of pepsin, rhizopuspepsin, and their active-site hydrogen bond mutants. The Journal of biological chemistry, 267(26), 18413–18418.
Munder, M. C., Midtvedt, D., Franzmann, T., Nüske, E., Otto, O., Herbig, M., Ulbricht, E., Müller, P., Taubenberger, A., Maharana, S., Malinovska, L., Richter, D., Guck, J., Zaburdaev, V., & Alberti, S. (2016). A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. eLife, 5, e09347. https://doi.org/10.7554/eLife.09347c
Politi, L., Chiancone, E., Giangiacomo, L., Cervoni, L., Scotto d'Abusco, A., Scorsino, S., & Scandurra, R. (2009). pH-, temperature- and ion-dependent oligomerization of Sulfolobus solfataricus recombinant amidase: a study with site-specific mutants. Archaea (Vancouver, B.C.), 2(4), 221–231. https://doi.org/10.1155/2009/280317
Rampelotto P. H. (2013). Extremophiles and extreme environments. Life (Basel, Switzerland), 3(3), 482–485. https://doi.org/10.3390/life3030482
Reed, C. J., Lewis, H., Trejo, E., Winston, V., & Evilia, C. (2013). Protein adaptations in archaeal extremophiles. Archaea (Vancouver, B.C.), 2013, 373275. https://doi.org/10.1155/2013/373275
The PyMOL Molecular Graphics System, Version 2.5.2 Schrödinger, LLC.
Theillet, F. X., Binolfi, A., Frembgen-Kesner, T., Hingorani, K., Sarkar, M., Kyne, C., Li, C., Crowley, P. B., Gierasch, L., Pielak, G. J., Elcock, A. H., Gershenson, A., & Selenko, P. (2014). Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chemical reviews, 114(13), 6661–6714. https://doi.org/10.1021/cr400695p
White, K. A., Grillo-Hill, B. K., & Barber, D. L. (2017). Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. Journal of cell science, 130(4), 663–669. https://doi.org/10.1242/jcs.195297
White, K. A., Grillo-Hill, B. K., Esquivel, M., Peralta, J., Bui, V. N., Chire, I., & Barber, D. L. (2018). β-Catenin is a pH sensor with decreased stability at higher intracellular pH. The Journal of cell biology, 217(11), 3965–3976. https://doi.org/10.1083/jcb.201712041
White KA, Kisor K, Barber DL. Intracellular pH dynamics and charge-changing somatic mutations in cancer. Cancer Metastasis Rev. 2019 Jun;38(1-2):17-24. doi: 10.1007/s10555-019-09791-8. PMID: 30982102.
White, K. A., Ruiz, D. G., Szpiech, Z. A., Strauli, N. B., Hernandez, R. D., Jacobson, M. P., & Barber, D. L. (2017). Cancer-associated arginine-to-histidine mutations confer a gain in pH sensing to mutant proteins. Science signaling, 10(495), eaam9931. https://doi.org/10.1126/scisignal.aam9931
Yao, X., Chen, C., Wang, Y., Dong, S., Liu, Y. J., Li, Y., Cui, Z., Gong, W., Perrett, S., Yao, L., Lamed, R., Bayer, E. A., Cui, Q., & Feng, Y. (2020). Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules. Science advances, 6(43), eabd7182. https://doi.org/10.1126/sciadv.abd7182

The protein streptavidin clamps onto the small molecule biotin in one of the tightest natural interactions ever discovered. Over the years, the strength of this...

Maybe you are about to start working with D-luciferin, and you begin reviewing protocols. But what you’re doing is a little different. Or what you...

Handles are common in everyday life. We use them to help us open doors, cabinets, or to hold coffee mugs, cookware, and tools. They are...

Labeling antibodies with biotin or a fluorophore enables scientists to detect, track, and quantify that antibody and the molecules that it binds to. We cover...
