DNA polymerase is an important and necessary reagent used every day in thousands of molecular biology laboratories around the
world. Aside from PCR, polymerases are used in a myriad of applications that require polymerases with specific abilities. There are many different polymerase enzymes available that exhibit different amplification abilities as well as various levels of fidelity (error rate) and processivity (number of bases a polymerase can add before disassociating from its substrate – template strand). Here, we provide you with a list of the relevant characteristics and functions of the most commonly used DNA polymerases to help you choose the right enzyme for your experiment.
For a quick comparison, scroll down or click here to view our chart of relevant characteristics of the polymerases discussed.
Most common polymerases
The first DNA polymerase, Pol I, was discovered in the late 1950s by Arthur Kornberg and his colleagues by purifying it from E. coli. Since then, many other polymerase enzymes have been discovered in bacteriophages or phages and engineered to produce amplicons for many downstream applications.
DNA Polymerase I
Pol I is made up of multiple domains that exhibit two different enzymatic activities: DNA-dependent DNA polymerase activity and exonuclease activity. The DNA polymerase activity achieves replication of DNA in a 5´ - 3´ direction and requires deoxyribo-nucleotides triphosphate (dNTPs) and a primer. On the other hand, the 3´ - 5´ exonuclease activity results in proofreading of the new DNA strand and removal of any mistakes. In addition, Pol I has a 5´ - 3´ exonuclease activity, which makes this enzyme useful in excision repair upstream of polymerization, removal of RNA primer, and nick translation. Pol I is also commonly used for 3´ DNA end removal or filling. Pol I’s molecular weight is 109 kDa and its optimal temperature is 37°C and can polymerize 1000 nucleotides per minute.
- 5´ - 3´ polymerase
- 3´ - 5´ exonuclease activity (proofreading)
- 109 kDa
- Optimal temperature is 37°C
- Polymerizes 1000 nucleotides per minute
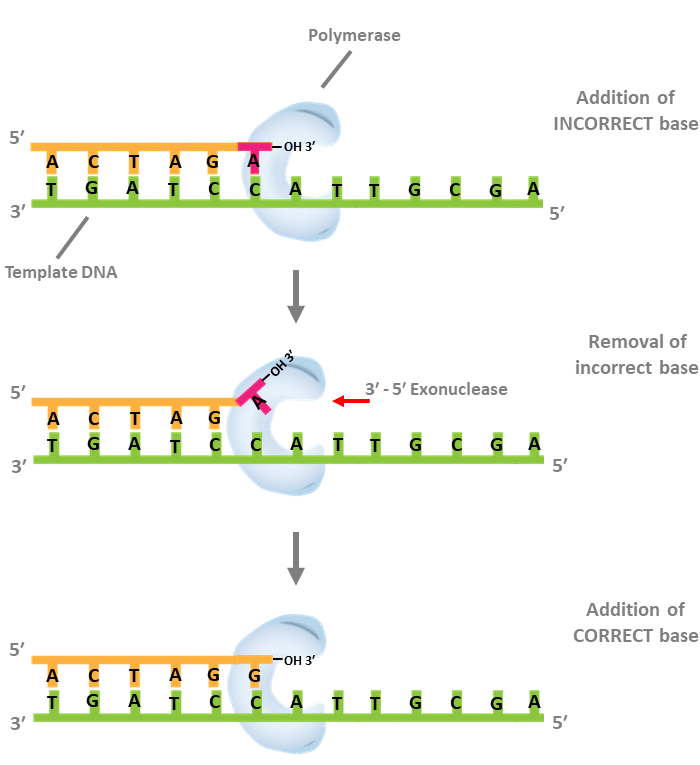
Proofreading (3´ - 5´ exonuclease) actvity
Klenow Fragment
The exonuclease activity of Pol I limits its use in molecular experiments because the 5´ - 3´ exonuclease activity might interfere with generating single-stranded DNA for downstream applications. Fortunately, researchers were able to separate these different functions by separating Pol I into two fragments. The small fragment contains the 5´ - 3´exonuclease activity while the large fragment or Klenow fragment retains the 5´ - 3´ polymerase and 3´ - 5´ proofreading activities. Thus, the Klenow fragment is very useful in double-stranded DNA synthesis (second strand cDNA synthesis), blunting by filling of 3´ ends (after restriction digestion or just filling in of large gaps), primer labeling (radiolabeled nucleotides onto 3’ ends), and DNA sequencing experiments.

Blunt ends and overhangs
It must be noted that both Pol I and Klenow fragments have an intermediate synthesis rate and low processivity (number of nucleotides incorporated before disassociation) and both result in a product with blunt ends. Error rates of DNA Pol I and Klenow fragment is (error/base pair/cycle) is 1 x 10-5 – 10-7.
- 5´ - 3´ polymerase
- 3´ - 5´ proofreading activity
- Intermediate synthesis rate
- Low processivity
- Blunt ended products
- Error rate is 1 x 10-5 – 10-7
T4 DNA Polymerase
In the presence of dNTPs, T4 polymerase synthesizes DNA strand in a 5´ - 3´ direction, while in the absence of dNTPs, it has 3´ - 5´ exonuclease activity. The DNA polymerase activity also requires a primer and single-stranded template. This enzyme is especially useful when trying to generate blunt ends and fill in gaps (fill in 5´ protruding ends). This enzyme can also be used for 3´ end labeling, nick translation and mutagenesis. Must be noted that T4 polymerase does not have 5´ - 3´ exonuclease activity. In addition, T4 DNA polymerase has an intermediate polymerase rate (~15,000 bases/minute), low processivity and an error rate of 1 x 10-6. This enzyme has a molecular weight of ~104 kDa, and optimal temperature is 37°C. It can be inactivated with EDTA or by incubation at 75°C.
- 5´ - 3´ polymerase
- 3´ - 5´ exonuclease activity
- Blunt ended products
- Low processivity
- Error rate of 1 x 10 -6
- Optimal temperature is 37°C
- Inactivated with EDTA or by incubation at 75°C
Taq Polymerase
One very distinctive characteristic of Taq polymerase is its thermostability. Taq polymerase, a 94 kDa enzyme, was first purified from Thermophilus aquaticus (Taq), a bacterium that thrives at high temperatures (70-80°C). Thus, Taq polymerase has a half-life of 40 minutes at 95°C making it useful for routine PCR, which uses high temperature for strand dissasociation. However, Taq does not have 3´ - 5´ exonuclease activity/proofreading; therefore, its high error rate (1 – 20 x 105) makes it a bad choice for applications requiring high fidelity (cloning, sequencing). The lack of this exonuclease activity does have some functionality since it results in single adenine overhang at the 3´ end (sticky ends), which can be ligated into plasmid with thymine 3´ overhangs (TA cloning).

TA Cloning
Newer Taq enzymes display high synthesis rate (1 kb per 30-60 seconds) and high processivity. In addition, Taq has 5´ - 3´ exonuclease activity, which can be used to generate a signal for amplification during PCR (qPCR). Also, Taq does require a primer, dNTPs, KCl and magnesium.
- Thermostable - ideal for routine PCR and TA cloning
- Requires a primer, dNTPs, KCl and magnesium
- NO 3´ - 5´ exonuclease activity/proofreading
- High error rate (1 – 20 x 10 5)
- High processivity
- Results in single adenine overhang at the 3´ end
Pfu Polymerase
Eventually, a thermostable 92 kDa polymerase with proofreading capabilities (3´ - 5´ exonuclease activity) was isolated from Pyrococcus furiosus Vc1 (Pfu). Pfu has a halflife of >120 minutes at 95°C and is the most accurate thermostable polymerase. Its proofreading activity allows Pfu to synthesize DNA strands with high fidelity with an error rate 1.6 x 10 -6 lower than Taq polymerase (1 error in 1.3 million base pairs). This processivity of Pfu is lower than that of Taq. Thus, this proofreading ability and low processivity results in a lower amplification rate than other polymerases because it takes about 2 minutes to amplify 1 kb at 70-75°C. Pfu’s proofreading does make it a good polymerase for mutation analysis, cloning, sequencing, and gene expression analysis. Successful PCR reactions with Pfu requires a primer, dNTPs, and magnesium and result in blunt ended product.
- Thermostable
- Proofreading capabilities (3´ - 5´ exonuclease activity)
- Most accurate thermostable polymerase
- Error rate is 1 x 10 -6 lower than Taq polymerase (1 error in 1.3 million base pairs)
- Low processivity
- Blunt ended products
Hot Start DNA Polymerase
Hot start method of PCR is a technique that is often used to improve PCR efficiency by preventing nonspecific product formation. This technique can involve excluding one or more reagents (such as the polymerase) until a specific temperature (60-80°C) is reached. In addition, hot start PCR could be done by reversibly inhibiting the polymerase.
Hot start Pfu or hot start Taq polymerases have been chemically modified to begin extending the DNA strand only after a specific high temperature is reached. This decreases the likelihood of nonspecific amplification at low temperatures.
Hot start PCR improves both multiplex and single-sided PCR reactions by suppressing these side reactions, primer oligomerization and mis-priming that reduce amplicon production. Hot start enzymes can be used for routine PCR, PCR involving low amount of template, and complex template. Hot start Pfu has 3´ - 5´ exonuclease (proofreading) making it a polymerase with high fidelity. Hot Start Taq has an error rate of 1 - 20 x 10-5, whereas Hot Start Pfu’s error rate is 1 - 2 x 10-6.
- Hot Start Pfu has 3´ - 5´ exonuclease
- Hot Start Taq has an error rate of 1 – 20 x 10 5
- Hot Start Pfu’s error rate is 1 - 2 x 10 -6
- High processivity
- Ideal for low template and complex template PCR
Find relevant characteristics of these polymerases in the table below:
DNA Polymerases
Additional Resources
- PCR Overview
- PCR Methods, Applications and Related GoldBio Products
- PCR & Gel Electrophoresis Troubleshooting Poster
- PCR Protocols
- PCR Master Mix Calculator
References
Alberts, B. M., & Frey, L. (1970). T4 Bacteriophage Gene 32: A Structural Protein in the Replication and Recombination of DNA. Nature, 227(5265), 1313-1318. doi:10.1038/2271313a0.
Bisen, P. (2014). Laboratory Protocols in Applied Life Sciences. doi:10.1201/b16575.
Cline, J. (1996). PCR fidelity of pfu DNA polymerase and other thermostable DNA polymerases. Nucleic Acids Research, 24(18), 3546-3551. doi:10.1093/nar/24.18.3546.
Holland, P. M., Abramson, R. D., Watson, R., & Gelfand, D. H. (1991). Detection of specific polymerase chain reaction product by utilizing the 5----3 exonuclease activity of Thermus aquaticus DNA polymerase. Proceedings of the National Academy of Sciences, 88(16), 7276-7280. doi:10.1073/pnas.88.16.7276.
Joyce, C. M., & Derbyshire, V. (1995). Purification of Escherichia coli DNA polymerase I and Klenow fragment. Methods in Enzymology DNA Replication, 3-13. doi:10.1016/0076-6879(95)62003-6.
Klenow, H., & Henningsen, I. (1970). Selective Elimination of the Exonuclease Activity of the Deoxyribonucleic Acid Polymerase from Escherichia coli B by Limited Proteolysis. Proceedings of the National Academy of Sciences, 65(1), 168-175. doi:10.1073/pnas.65.1.168.
Kucera, R. B., & Nichols, N. M. (2008). DNA-Dependent DNA Polymerases. Current Protocols in Molecular Biology. doi:10.1002/0471142727.mb0305s84.
Lundberg, K. S., Shoemaker, D. D., Adams, M. W., Short, J. M., Sorge, J. A., & Mathur, E. J. (1991). High-fidelity amplification using a thermostable DNA polymerase isolated from Pyrococcus furiosus. Gene, 108(1), 1-6. doi:10.1016/0378-1119(91)90480-y.
Mcinerney, P., Adams, P., & Hadi, M. Z. (2014). Error Rate Comparison during Polymerase Chain Reaction by DNA Polymerase. Molecular Biology International, 2014, 1-8. doi:10.1155/2014/287430.
Rittié, L., & Perbal, B. (2008). Enzymes used in molecular biology: A useful guide. Journal of Cell Communication and Signaling, 2(1-2), 25-45. doi:10.1007/s12079-008-0026-2.
Śpibida, M., Krawczyk, B., Olszewski, M., & Kur, J. (2016). Modified DNA polymerases for PCR troubleshooting. Journal of Applied Genetics, 58(1), 133-142. doi:10.1007/s13353-016-0371-4.
Steitz, T. A. (1999). DNA Polymerases: Structural Diversity and Common Mechanisms. Journal of Biological Chemistry, 274(25), 17395-17398. doi:10.1074/jbc.274.25.17395.
Fernanda Ruiz is a science content writer at Gold Biotechnology. She holds a bachelor's of science in biology from St. Mary's University and a PhD in molecular biology from Baylor College of Medicine.
https://www.goldbio.com/pcr-handbook-download-for-...






