Why Lysozyme and Zymolyase are Important in Cell Lysis Buffers
by Pallabi Roy Chakravarty, Ph.D.

by Pallabi Roy Chakravarty, Ph.D.
Lysozyme and zymolyase are enzymes that degrade cell walls of bacteria and yeast cells. When bacterial or yeast cells are lysed during experiments, these enzymes are added in cell lysis buffers to break down the cell wall. Each has properties making them ideal for different procedures.
In many gene cloning experiments, the cloning host is either a bacterium or yeast. Also, very often, these microorganisms serve as the expression host. So, in these cases, you would need to lyse the host cells to extract your cloned DNA or the resulting recombinant product molecule. For this purpose, lysozyme and zymolyase are very helpful.
The buffer used for breaking down the host cells to extract your target molecules is technically called a “lysis buffer.” Here is an article with a lot of important information and tips about cell lysis buffers and the common chemicals these buffers contain.
Because of lysozyme’s and zymolyase’s unique and specific activities toward bacterial and yeast cell walls, respectively, they are specifically added when you want to break open microbial cells.
In this article, we will delve into these enzymes, and explore the biochemistry behind how they degrade cell walls, helping with cell lysis in the process.
And to put things more into perspective, we will also talk about some excellent resources that GoldBio offers you in this context – which may make your microbial lysis experiments so much easier.
Bacterial cell lysis kits to the rescue
Lysozyme is an enzyme of the type glycoside hydrolase. It breaks down peptidoglycan, a primary constituent of Gram-positive bacterial cell walls. Lysozyme is used in cell lysis buffers when the target cells are bacterial.
Lysozyme is a potent lytic agent of bacterial cell walls. This is evident by the fact that it is found in human fluids such as saliva and tears as a protective chemical against pathogenic bacteria. Scientists have taken advantage of this property when developing methods for bacterial cell lysis.
Gram-positive bacteria have cell walls composed of peptidoglycan. Peptidoglycan is a polymer of NAM (N-acetyl muramic acid) and NAG (N-acetyl glucosamine), cross-linked with a short chain of polypeptides.
Lysozyme breaks the bond between NAG and NAM molecules by a hydrolysis reaction.
Figure 1. Lysozyme catalyzing the breakdown of the
peptidoglycan layer.
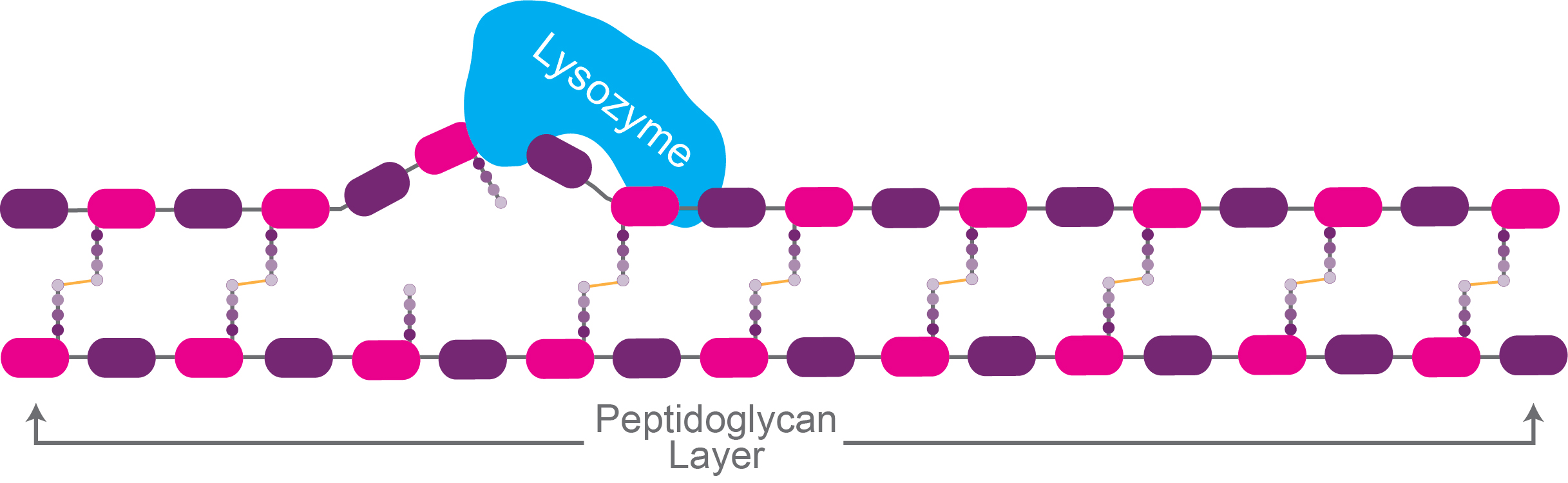
Figure 1. Lysozyme catalyzing the breakdown of the
peptidoglycan layer.

After being treated with lysozyme, or a similar enzyme, a cell is stripped of its cell wall, and at this stage it is technically known as a “spheroplast.”
Lysozyme is very helpful during cell lysis when using Gram-positive organisms in your experiment. However, when you are working with Gram-negative bacteria, the situation gets trickier. And most often, we work with E. coli, which is Gram-negative.
Gram-negative bacterial cells have a lipopolysaccharide
(LPS) layer that surrounds the peptidoglycan. Because of this, lysozyme cannot
effectively reach and break down the peptidoglycan in the cell wall of
Gram-negative bacterial cells.
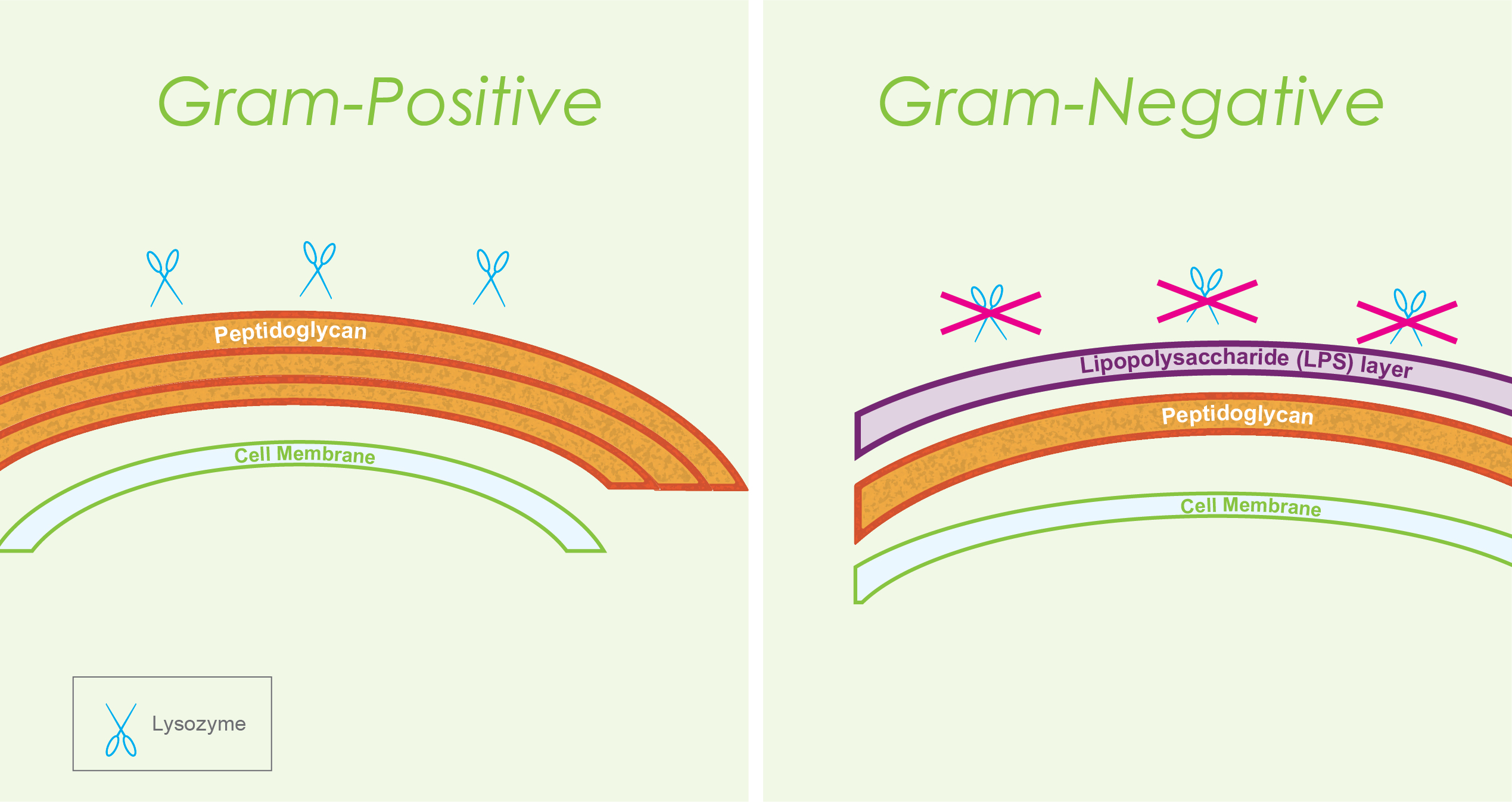
Figure 3. Difference between Gram-positive and
Gram-negative cell walls. This difference accounts for the fact that lysozyme
is effective in breaking open Gram-positive bacterial cells, but not so much
with lysing Gram-negative bacteria.
The way to break the LPS layer is by treating the cells with detergents. However, when you want to extract proteins in their native (non-denatured) structure, using denaturing detergents such as SDS might become damaging. If you want to know more about this, here are some great insights on how SDS denatures proteins.
To get around this issue means that you’d need to strike a balance between being able to disrupt the LPS layer while not denaturing the cellular proteins that you are extracting. For this, you’d need to have a non-denaturing detergent in your cell lysis buffer.
A great example of this is our Bacterial Cell Lysis kit. The mild detergent NP-40 in this buffer breaks the LPS and also the bacterial cell membranes but does not denature proteins.
Further, it contains salts and tris to maintain the optimal pH and ionic balance required for cell lysis. The buffer kit provides a tube of lysozyme in case you want to add it in your lysis buffer. It also has nucleases to degrade nucleic acids during cell lysis so that you can extract proteins with minimal nucleic acid contamination.
Depending on what you need in your experiment, you may consider adding protease inhibitors, phosphatase inhibitors and denaturants into this buffer for cell lysis. Also, EDTA is a common chemical in bacterial lysis buffers. By chelating divalent cations, it modifies the surface charge of bacterial cell surfaces and helps expose the peptidoglycan layer to lysozyme for cleavage.
Very similar to the Bacterial Cell Lysis kit is the mammalian cell lysis buffer kit and the tissue cell lysis buffer kit. Animal cells do not have peptidoglycan surrounding them, and they do not have cell walls – so it is much easier to lyse them compared to bacterial cells.
Buffers for animal cell lysis just have buffering agents such as tris, salts for ionic balance and mild detergents – especially if you want to perform cell lysis to extract proteins in their native structures.
However, when you are working with yeast cells that have strong cell walls, the path is not as smooth as what it is while lysing animal cells. The way around this is using the enzyme zymolyase which makes it easier to lyse yeast cell walls.
Zymolyase is an enzyme that helps break down the cell walls of yeast cells. This enzyme uses β-1,3-glucan laminaripentaohydrolase and β-1,3-glucanase activities in its function to lyse yeast cell walls.
Zymolyase breaks the β-1,3-bonds joining glucose residues in the yeast cell wall, producing laminaripentaose, and lysing the cell wall polymeric structures.
Since the yeast strain Saccharomyces cerevisiae is widely used in lab experiments, zymolyase becomes a very handy reagent. Like bacterial cells, once the cell wall is removed, yeast cells become spheroplasts.
We have an excellent cell lysis kit specifically for yeast cells, which comes with zymolyase. The lysis buffer has other usual components to maintain ionic strength and pH, and non-ionic mild detergents for helping with membrane lysis. And you can optimize it with protease inhibitors, chelators, and reducing agents, depending on your experimental needs.
In this article, we covered why lysozyme and zymolyase are so useful to add in cell lysis buffers in experiments that involve breaking down microbial cells. These along with other enzymes and reagents optimize the entire process, ultimately leading to good yields for your work.
Dean and Ward. 1992. The use of EDTA or polymyxin with lysozyme for the recovery of intracellular products from Escherichia coli. Biotechnology Techniques. Volume 6, pages 133-138.
Falgenhauer et al. 2021. Evaluation of an E. coli Cell Extract Prepared by Lysozyme‐Assisted Sonication via Gene Expression, Phage Assembly and Proteomics. Chembiochem. 22(18): 2805–2813
Rodríguez-Peña et al. 2013. Activation of the yeast cell wall integrity MAPK pathway by zymolyase depends on protease and glucanase activities and requires the mucin-like protein Hkr1 but not Msb2. FEBS Letters. Volume 587, Issue 22, 15 November 2013, Pages 3675-3680
Shehadul Islam et al. 2017. A Review on Macroscale and Microscale Cell Lysis Methods. Micromachines. 8(3):83

Making IPTG stock solution involves weighing out IPTG powder, pouring it into a conical tube or cylinder, and adding deionized water to the desired volume....

IPTG and auto-induction are two ways to induce protein expression in bacteria. They work similarly, but have different trade-offs in terms of convenience. While IPTG...

The final concentration of IPTG used for induction varies from 0.1 to 1.0 mM, with 0.5 or 1.0 mM most frequently used. For proteins with...

A His-tag is a stretch of 6-10 histidine amino acids in a row that is used for affinity purification, protein detection, and biochemical assays. His-tags...
