How Do Bacteria Develop Antibiotic Resistance?
by Tyasning Kroemer, Ph.D.

by Tyasning Kroemer, Ph.D.
An antibiotic is an antimicrobial chemical that kills or inhibits the growth of microorganisms. Antibiotics have been used for a long time to cure many deadly infections, such as bubonic plaque, diphtheria, and tuberculosis.
The mechanisms of action of antibiotics depends on their structures and their affinity level with the target in the microorganism cells.
Mechanisms by which antibiotics target bacteria are:

After the first discovery of an antibiotic, penicillin, in 1928 by Alexander Fleming, many antibiotics have been isolated and used for the treatment of many diseases. Unfortunately, many bacterial strains have developed resistance against widely used antibiotics.
Ampicillin is an antibiotic with a β-lactam ring that inhibits penicillin-binding proteins (PBPs), involved in peptidoglycan biosynthesis. When the PBPs interacts with β-lactam rings, these proteins can’t catalyze the synthesis of new peptidoglycan, disrupting the formation of the bacterial cell wall.
Bacteria become resistant to ampicillin by producing β-lactamase enzyme. This enzyme cleaves the β-lactam ring of ampicillin to inactivate it. Many cloning vectors with a resistance gene, bla gene, produce β-lactamase enzyme.
Chloramphenicol is a broad-spectrum antibiotic, targeting a wide range of microorganisms. This antibiotic inhibits the activity of ribosomal peptidyl transferase. As a result, it inhibits protein synthesis.
Bacteria develop chloramphenicol resistance by using the activity of chloramphenicol acetyl transferase (CAT) to modify chloramphenicol.

Examples of Different Mechanisms of Antibiotic Resistance. 1) Antibiotic degradation by β-lactamases. 2) Antibiotic modification by chloramphenicol acetyl transferase (CAT) and amino-phospotransferase (APH). 3) Antibiotic efflux by Tet proteins. 4) Ribosome protection by Tet proteins.
Kanamycin is a member of aminoglycoside family of antibiotics. This antibiotic interacts with three ribosomal proteins and rRNA in the 30S ribosomal subunit, causing the inhibition of protein synthesis.
Resistance to kanamycin involves the activity of amino-phosphotransferase enzymes.To inactivate and modify kanamycin, an amino-phosphotransferase transfers a phosphate group from their ATP (Adenine Triphosphate) to kanamycin. Two examples of these enzymes are Aph (3’)-I and Aph (3’)-II.
Tetracyclines bind to a site on the 30S ribosomal subunit to prevent the attachment of aminoacyl tRNA to the acceptor site and inhibit the synthesis of protein.
The resistance against tetracyclines involves Tet proteins. Tet proteins catalyze the antibiotic efflux (by pumping tetracyclines out of the cell) or protect the ribosome (by blocking tetracyclines from binding to the ribosome). Tet(A), Tet(B), Tet(C), Tet(D), Tet(E), Tet(G) and Tet(H) are efflux proteins. Tet(M) and Tet(O) are examples of proteins involved in ribosomal protection.
Bacteria can obtain antibiotic resistance from:
The resistance gene can produce:
The exchange or horizontal gene transfer (HGT) of resistance genes by bacteria uses three main strategies:

Bacteria that can’t get antibiotic resistance naturally will not survive and grow in the growth medium containing antibiotics. Based on this, molecular biologists design special plasmid vectors.
These vector tools carry a specific DNA insert that the researcher want to amplify and an antibiotic resistance gene as a selectable marker. Competent bacteria cells will take up these plasmid vectors inside the cells through artificial transformation (by heat shock or electroporation).

After artificial transformation and antibiotic selection, only bacteria containing the plasmid vector with a resistance gene survive in the cell culture. During antibiotic selection, antibiotics kill bacteria that don’t have the plasmid vector with the resistance gene in the cell.
Below are common problems during antibiotic selection and a simple troubleshooting guide for these problems:
Competent Cells
GB10B™ Chemically Competent E. coli Cells (Catalog No. CC-100)
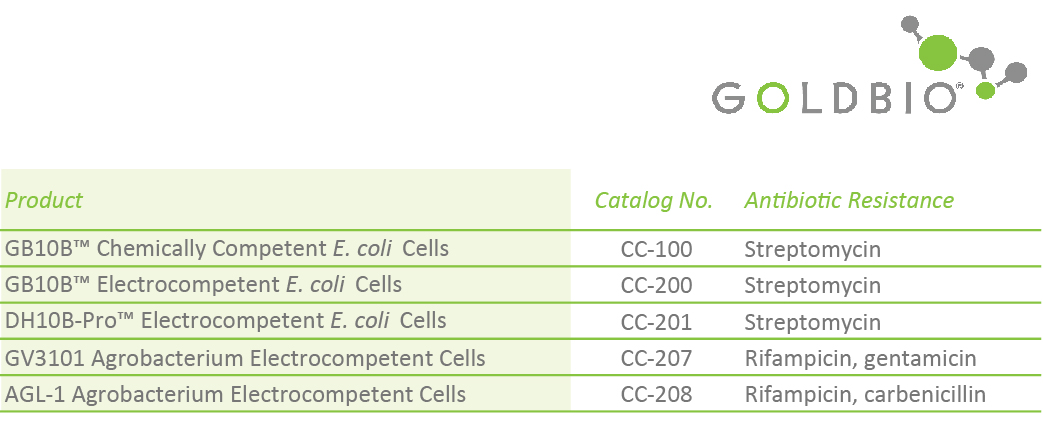
GB10B™ Electrocompetent E. coli Cells (Catalog No. CC-200)
GB5-alpha™ Chemically Competent E. coli Cells (Catalog No. CC-101)
BL21 (DE3) Chemically Competent E. coli Cells (Catalog No. CC. 103)
DL39 (DE3) Chemically Competent E. coli Cells (Catalog No. CC-104)
GB10B-Pro™ Electrocompetent E. coli Cells (Catalog No. CC-201)
GB5-alpha™ Electrocompetent E. coli Cells (Catalog No. CC-203)
BL21 (DE3) Electrocompetent E. coli Cells (Catalog No. CC-204
GV3101 Agrobacterium Electrocompetent Cells (Catalog No. CC-207)
AGL-1 Agrobacterium Electrocompetent Cells (Catalog No. CC-208)
Popular Antibiotics Products
Ampicillin (Sodium), USP Grade (Catalog No. A-301)
Chloramphenicol, USP Grade (Catalog No. C-105)
Kanamycin Monosulfate, USP Grade (Catalog No. K-120)
Tetracycline Hydrochloride (Catalog No. T-101)
Carbenicillin (Disodium), USP Grade (Catalog No. C-103)
Penicillin G Sodium Salt, USP Grade (Catalog No. P-304)
Penicillin G Potassium Salt, USP Grade (Catalog No. P-303)
Ampicillin (Sodium) EZ Pak™ for 100 mg/mL Solution (Catalog No. A-301-EZ)
Streptomycin Sulfate EZ Pak™ for 100 mg/mL Solution (Catalog No. S-150-EZ)
Carbenicillin (Disodium) EZ Pak™ for 100 mg/mL Solution (Catalog No. C-103-EZ)
Kanamycin Monosulfate EZ Pak™ for 50 mg/mL Solution (Catalog No. K-120-EZ)
And many more !
Bennett, P. M. (2008). Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. British Journal of Pharmacology, 153(S1), S347-S357. doi:10.1038/sj.bjp.0707607
Demain, A. L., & Elander, R. P. (1999). The β-lactam antibiotics: past, present, and future. Antonie van Leeuwenhoek, 75(1), 5-19. doi:10.1023/A:1001738823146
Garneau-Tsodikova, S., & Labby, K. J. (2016). Mechanisms of Resistance to Aminoglycoside Antibiotics: Overview and Perspectives. MedChemComm, 7(1), 11-27. doi:10.1039/C5MD00344J
Gould, K. (2016). Antibiotics: from prehistory to the present day. Journal of Antimicrobial Chemotherapy, 71(3), 572-575. doi:10.1093/jac/dkv484
Kapoor, G., Saigal, S., & Elongavan, A. (2017). Action and resistance mechanisms of antibiotics: A guide for clinicians. Journal of anaesthesiology, clinical pharmacology, 33(3), 300.
Kohanski, M. A., Dwyer, D. J., & Collins, J. J. (2010). How antibiotics kill bacteria: from targets to networks. Nature reviews. Microbiology, 8(6), 423-435. doi:10.1038/nrmicro2333
Martinez, J. L., & Baquero, F. (2000). Mutation Frequencies and Antibiotic Resistance. Antimicrobial Agents and Chemotherapy, 44(7), 1771-1777. doi:10.1128/aac.44.7.1771-1777.2000
Munita, J. M., & Arias, C. A. (2016). Mechanisms of Antibiotic Resistance. Microbiology spectrum, 4(2), 10.1128/microbiolspec.VMBF-0016-2015. doi:10.1128/microbiolspec.VMBF-0016-2015
Nelson, M. L., Dinardo, A., Hochberg, J., & Armelagos, G. J. (2010). Brief communication: mass spectroscopic characterization of tetracycline in the skeletal remains of an ancient population from Sudanese Nubia 350–550 CE. American journal of physical anthropology, 143(1), 151-154.
Peterson, E., & Kaur, P. (2018). Antibiotic Resistance Mechanisms in Bacteria: Relationships Between Resistance Determinants of Antibiotic Producers, Environmental Bacteria, and Clinical Pathogens. Frontiers in Microbiology, 9(2928). doi:10.3389/fmicb.2018.02928
Preston, A. (2003). Choosing a cloning vector. In E. coli Plasmid Vectors (pp. 19-26): Springer.
Reece, K. S., & Phillips, G. J. (1995). New plasmids carrying antibiotic-resistance cassettes. Gene, 165(1), 141-142. doi:https://doi.org/10.1016/0378-1119(95)00529-F
Rolinson, G., Macdonald, A., & Wilson, D. (1977). Bactericidal action of β-lactam antibiotics on Escherichia coli with particular reference to ampicillin and amoxycillin. Journal of Antimicrobial Chemotherapy, 3(6), 541-553.
Rossolini, G., Arena, F., & Giani, T. (2017). Mechanisms of Antibacterial Resistance. In (pp. 1181-1196.e1181).
Sharpe, G. S. (1984). Broad host range cloning vectors for Gram-negative bacteria. Gene, 29(1), 93-102. doi:https://doi.org/10.1016/0378-1119(84)90170-7
Speer, B. S., Shoemaker, N. B., & Salyers, A. A. (1992). Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clinical microbiology reviews, 5(4), 387-399. doi:10.1128/cmr.5.4.387
Woodford, N., & Ellington, M. J. (2007). The emergence of antibiotic resistance by mutation. Clinical Microbiology and Infection, 13(1), 5-18

A His-tag is a stretch of 6-10 histidine amino acids in a row that is used for affinity purification, protein detection, and biochemical assays. His-tags...

Competent cells such as DH5a, DH10B, and BL21 will maintain their transformation efficiency for at least a year with proper storage. It is important to...

Ni2+ ions give nickel agarose beads their characteristic blue color. This blue color can fade or disappear completely when loading his-tagged proteins onto the column....

Nickel agarose beads change from blue to a brown or black color when the nickel ions have been reduced from a Ni2+ to a Ni1+...
