Picture yourself staying a winter night in the middle of the Arctic or lying in boiling thermal water with no protection or shelter.
It’s not possible – not for us anyway. However, it can be entirely possible for certain microbes to spend their lives in extreme environments.
Microbes have inhabited Earth for millions and millions of years and have evolved even to survive in very inhospitable environments.
An extreme microbiome is composed of microorganisms (extremophiles) able to survive under harsh conditions where no other living being would have any chance of surviving.
The extreme environments where these microbes live are characterized by low temperature, high temperatures, high salt content or hypersaline, and high/low pH, which are distributed worldwide.
These extreme environments actually retain a high biological diversity which has been approached to create biotechnological tools.
One of the most famous tools was Taq polymerase obtained from the bacterium Thermus aquaticus for thermal environments.
Many other microorganisms are being explored to speed their use for future needs in different industries such as chemical, food, and pharmaceuticals.
In celebration of these extreme microbes, we’re taking you through some of the most extreme environments and taking a look at bacterial distribution.
In this article:
Hypersaline microbiomes
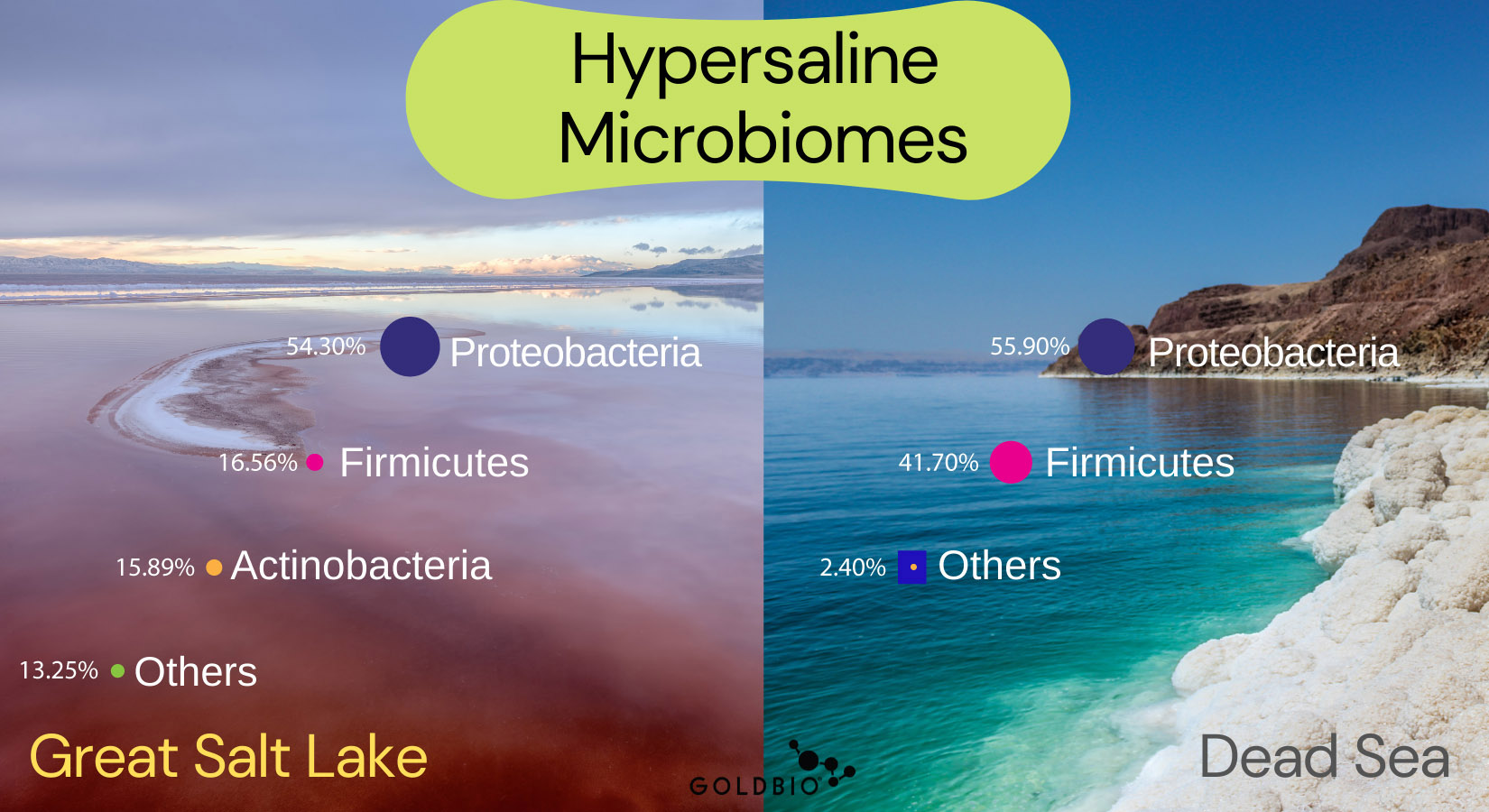
The microbes living in a hypersaline environment are called halophiles. Places such as the Dead Sea or The Great Salt Lake have allowed researchers to really study halophiles and the main taxa predominating these environments.
For instance, in the Great Salt Lake in Utah, USA, bacteria like Halobacterium and Halococcus are abundant.
Interestingly, these bacteria depend on algae to get their nutrients. They use organic matter produced by the algae, and the algae uses ammonia produced by the bacteria.
Furthermore, another recent study reported that the Great Salt Lake is characterized by Proteobacteria (54.3%), Firmicutes (16.56%), and Actinobacteria (15.89%) phyla with the remaining 14% as other unknown microbes (Bhattarai et al. 2021).
Usually, the Great Salt Lake is compared to the Dead Sea located in the western part of Jordan.
The Dead Sea is characterized by 52% Archea and 45% Bacteria. Within Bacteria, Proteobacteria (55.9%) and Firmicutes (41.7%) are the most abundant in Dead Sea.
Concerning dominant bacterial genera, two species were found as predominant: Acinetobacter (45%) and Bacillus (35%).
Some other factors explaining why these two salty places present different microbiomes include the temperature ranges and sea level.
On one side, the range of temperatures in the Great Salt Lake goes from -5° up to 35°C while the Dead Sea usually ranges between 21° to 36°C.
Furthermore, the Dead Sea is considered the Earth's lowest elevation on land (more than 420 m below sea level).
Thermal microbiomes

Microbes that can survive in very hot temperatures are called thermophiles.
A study at Yellowstone National Park characterized by waters between 44–75°C showed variations in the resident bacterial population over three years of study at the three sites with different temperatures.
For instance, the dominant bacterial community changed from moderately thermophilic to photosynthetic members (Cyanobacteria and Chloroflexi) at 44°C.
At 63 °C, researchers found a bacterial community of thermophilic bacteria that are also photosynthetic, or photosynthetic-thermophiles (Deinococcus-Thermus).
At 75°C, the bacterial community was mostly made up of bacteria that were not photosynthetic-thermophiles (Bowen De Leon et al., 2013). Interestingly, a high proportion of Chloroflexi (~17.8%), Bacteroidetes (~17.7%), Proteobacteria (~13.5%), and Firmicutes (~12.0%) was found in Yellowstone hot springs (Jiang et al. 2017).
In another study based on 16S rRNA sequencing technology from Sungai Klah hot spring in Malaysia (50–110°C and pH 7–9), Firmicutes (37.15%) and Proteobacteria (19.26%) were found to be the most abundant phyla. Here the thermophile Hydrogenobacter spp. represented one of the major taxa.
Researchers believe this diversified phylogenetic community is due to the composition of its natural environment. For instance, the pile of fallen leaves in the ground (also called plant litter), plus shallow streams with their broad temperature and pH ranges, both contribute to this diversity. Furthermore, plant litter and geochemical parameters are also associated with allowing symbiotic relationships to occur between the members of the thermal community (Chan et al. 2015).
Permafrost microbiomes

Microbes surviving very low temperatures are called psychrophiles. Permafrost environments are characterized by a permanent ice layer on their soils.
The Arctic and Antarctic are two of the most geographically distant bioregions recognized as permafrost environments. However, other conditions such as low temperature, low availability of carbon sources, and extreme seasonality in light conditions also characterize the Arctic and Antarctic.
Independent studies have been carried out in both regions.
The most dominant bacterial phyla detected across all permafrost soil samples in the Arctic were Actinobacteria, (24.62%), followed by Proteobacteria (15.37%), Acidobacteria (10.29%), Firmicutes (4.25%), others (45.47%) (Tripathi et al. 2019).
In contrast, studies in the Antarctic have been focused on marine sediments. Gammaproteobacteria (92.4%), followed by Alphaproteobacteria (2.5%), Firmicutes (1.5%), Bacteroidetes (1.1%), others (1.5%) were found as the most dominant phyla (Franco et al 2017).
Actinobacteria was characteristic of Artic, while Gammaprotebacteria was more characteristic of the Antarctic.
It has been suggested that Actinobacteria are successful in colonizing permafrost soils because they have adapted to harsh abiotic conditions and can degrade complex organic compounds, such as cellulose and lignin.
The hypothesis of how these microbes survive in these environments has been studied. Research suggests diverse bacteria can assimilate carbon dioxide from the air to create feed products using atmospheric trace gases like H2 and CO as energy resources (Lambrechts et al., 2019).
Gammaproteobacteria is also frequently isolated from extreme environments, including cold habitats and deep-sea sediments. These bacteria are able to produce n-3 poly-unsaturated fatty acid (3-PUFAs), which modify the lipid membranes make them more flexible to survive under cold environments (Parrilli et al., 2019).
Desert microbiomes (hot deserts)

We’ve already talked about the thermophiles, microbes surviving hot environments.
Desert environments are characterized by high salinity, drought, and extreme temperatures.
Incredibly, deserts are found on every continent, making up 33% of the total land area worldwide (Alsharif et al., 2020).

Desert soils from across the world typically contain many ubiquitous phyla, including Actinobacteria, Bacteroidetes, and Proteobacteria (Makhalanyane et al 2015).
In the Sahara Desert of Egypt, bacterial communities of Proteobacteria (46.0%, predominantly Ochrobactrum), Actinobacteria (20.7%, predominantly Rhodococcus), and Firmicutes (11.3%, predominantly Bacillus) were the dominant phyla (Belov et al 2018).
However, in the Atacama Desert of Chile, most research takes model plants species and studies their rhizosphere-associated microbiomes.
The rhizosphere is the narrow region of soil or substrate directly influenced by root secretions and associated soil microorganisms.

While the study of the rhizosphere can give us a very narrow idea of a specific desert soil, these microbes populations can be compared among plant species from different deserts.
For instance, Araya et al. (2020) studied Cistanthe longiscapa bloom (Montiaceae family). They found Actinobacteria (43.4%) and Proteobacteria (20.7%) as the most abundant phyla followed by Chloroflexi (8.9%), Firmicutes (6.9%), Bacteroidetes (6.5%), Acidobacteria (4.5%), Planctomycetes (3.5%) and Gemmatimonadetes (2.1%).
Furthermore, Fuentes et al., 2020 investigated the rhizosphere of Baccharis scandens and Solanum chilense, two native plants from the Atacama Desert. He also found Actinobacteria and Proteobacteria as the dominant taxa in the rhizosphere of both plants.
Actinobacteria often prevail in desert soil phylogenetic surveys.
Evidence that actinobacteria is the dominant phylum in arid environments is perhaps unsurprising, given their wide metabolic and degradative capacity, competitive advantages via secondary metabolite synthesis, and multiple UV repair mechanisms as UV light which hampers DNA molecules (Makhalanyane et al. 2015).
Volcanic microbiomes

Volcanic environments are particularly challenging as they encompass several extreme characteristics, such as high temperatures (>70 °C), heavy metals (Cr, Cd, Cu), and particularly high acidity (pH < 4) (Peña-Ocaña et al. 2021).
The active Poás Volcano, Laguna Caliente, in Costa Rica was sampled. This volcano is characterized by near-ambient to boiling temperatures, frequent internal eruptions, and pH fluctuations from −0.87 to 1.5.
Indeed, this volcano is an ultra-acidic, sulfurous crater lake primarily sourced through a subsurface groundwater system.
Researchers found that at the lake bottom, there was extremely low biodiversity of microorganisms dominated by Acidiphilium spp. belonging to Proteobacteria (Wang et al. 2022).
On the other hand, one of the most active volcanoes in Mexico is El Chichón volcano. The authors found that the bacteria phyla Firmicutes (52.7%, mostly Alicyclobacillus and Sulfobacillus) and Proteobacteria (44.8%, mostly Bradyrhizobium, Methylobacterium, Sediminibacterium) were the most abundant at 92°C (Peña-Ocaña et al., 2021).
Conclusion
Environments that are hostile to humans have been described as extreme. Microbes have evolved an arsenal of metabolites and enzymes to survive and even thrive in extremely challenging conditions over the years.
In this respect, the study of extremophiles and their cell components makes them suitable candidates for screening potentially interesting enzymes and novel compounds that provide benefits in chemical, food, pharmaceutical, paper, textile industries, and environmental biotechnology.
Keywords
Extremophiles, extreme environments, microbiome.
References
Aburto-Medina, A., Shahsavari, E., Cohen, M., Mantri, N., & Ball, A. S. (2020). Analysis of the microbiome (Bathing Biome) in geothermal waters from an Australian balneotherapy centre. Water (Switzerland), 12(6). https://doi.org/10.3390/W12061705
Alsharif, W., Saad, M. M., & Hirt, H. (2020). Desert Microbes for Boosting Sustainable Agriculture in Extreme Environments. Frontiers in Microbiology, 11(July). https://doi.org/10.3389/fmicb.2020.01666
Araya, J. P., González, M., Cardinale, M., Schnell, S., & Stoll, A. (2020). Microbiome Dynamics Associated With the Atacama Flowering Desert. Frontiers in Microbiology, 10(January), 1–13. https://doi.org/10.3389/fmicb.2019.03160
Bhattarai, B., Bhattacharjee, A. S., Coutinho, F. H., & Goel, R. K. (2021). Viruses and Their Interactions With Bacteria and Archaea of Hypersaline Great Salt Lake. Frontiers in Microbiology, 12(September). https://doi.org/10.3389/fmicb.2021.701414
Baxter, B. K. (2018). Great Salt Lake microbiology: a historical perspective. International Microbiology, 21(3), 79–95. https://doi.org/10.1007/s10123-018-0008-z
Belov, A. A., Cheptsov, V. S., & Vorobyova, E. A. (2018). Soil bacterial communities of sahara and gibson deserts: Physiological and taxonomical characteristics. AIMS Microbiology, 4(4), 685–710. https://doi.org/10.3934/microbiol.2018.4.685
Cao, S., Zhang, W., Ding, W., Wang, M., Fan, S., Yang, B., McMinn, A., Wang, M., Xie, B. Bin, Qin, Q. L., Chen, X. L., He, J., & Zhang, Y. Z. (2020). Structure and function of the Arctic and Antarctic marine microbiota as revealed by metagenomics (Microbiome (2020) 8 (47) DOI: 10.1186/s40168-020-00826-9). Microbiome, 8(1), 1–12. https://doi.org/10.1186/s40168-020-00871-4
Chan, C. S., Chan, K. G., Tay, Y. L., Chua, Y. H., & Goh, K. M. (2015). Diversity of thermophiles in a Malaysian hot spring determined using 16S rRNA and shotgun metagenome sequencing. Frontiers in Microbiology, 6(MAR), 1–15. https://doi.org/10.3389/fmicb.2015.00177
De León, K. B., Gerlach, R., Peyton, B. M., & Fields, M. W. (2013). Archaeal and bacterial communities in three alkaline hot springs in heart lake geyser basin, yellowstone national park. Frontiers in Microbiology, 4(NOV), 1–10. https://doi.org/10.3389/fmicb.2013.00330
Franco DC, Signori CN, Duarte RTD, Nakayama CR, Campos LS and Pellizari VH (2017). High Prevalence of Gammaproteobacteria in the Sediments of Admiralty Bay and North Bransfield Basin, Northwestern Antarctic Peninsula. Front. Microbiol. 8:153. doi: 10.3389/fmicb.2017.00153
Fuentes, A., Herrera, H., Charles, T. C., & Arriagada, C. (2020). Fungal and bacterial microbiome associated with the rhizosphere of native plants from the atacama desert. Microorganisms, 8(2). https://doi.org/10.3390/microorganisms8020209
Jacob, J. H., Hussein, E. I., Shakhatreh, M. A. K., & Cornelison, C. T. (2017). Microbial community analysis of the hypersaline water of the Dead Sea using high-throughput amplicon sequencing. MicrobiologyOpen, 6(5), 1–6. https://doi.org/10.1002/mbo3.500
Jiang, X., & Takacs-Vesbach, C. D. (2017). Microbial community analysis of pH 4 thermal springs in Yellowstone National Park. Extremophiles, 21(1), 135–152. https://doi.org/10.1007/s00792-016-0889-8
Jones, D. L., & Baxter, B. K. (2017). DNA repair and photoprotection: Mechanisms of overcoming environmental ultraviolet radiation exposure in halophilic archaea. Frontiers in Microbiology, 8(SEP), 1–16. https://doi.org/10.3389/fmicb.2017.01882
Lambrechts, S., Willems, A., & Tahon, G. (2019). Uncovering the uncultivated majority in antarctic soils: Toward a synergistic approach. Frontiers in Microbiology, 10(FEB), 1–19. https://doi.org/10.3389/fmicb.2019.00242
Makhalanyane, T. P., Valverde, A., Gunnigle, E., Frossard, A., Ramond, J. B., & Cowan, D. A. (2015). Microbial ecology of hot desert edaphic systems. FEMS Microbiology Reviews, 39(2), 203–221. https://doi.org/10.1093/femsre/fuu011
Mehta, P., Yadav, M., Ahmed, V., Goyal, K., Pandey, R., & Chauhan, N. S. (2021). Culture-Independent Exploration of the Hypersaline Ecosystem Indicates the Environment-Specific Microbiome Evolution. Frontiers in Microbiology, 12(October). https://doi.org/10.3389/fmicb.2021.686549.
Parrilli E, et al. (2019). The art of adapting to extreme environments: The model system Pseudoalteromonas. Phys Life Rev, https://doi.org/10.1016/j.plrev.2019.04.003
Peña-Ocaña, B. A., Ovando-Ovando, C. I., Puente-Sánchez, F., Tamames, J., Servín-Garcidueñas, L. E., González-Toril, E., Gutiérrez-Sarmiento, W., Jasso-Chávez, R., & Ruíz-Valdiviezo, V. M. (2022). Metagenomic and metabolic analyses of poly-extreme microbiome from an active crater volcano lake. Environmental Research, 203(April 2021). https://doi.org/10.1016/j.envres.2021.111862
Post, F. J. (1977). The microbial ecology of the Great Salt Lake. Microbial Ecology, 3(2), 143–165. https://doi.org/10.1007/BF02010403
Sousa, A. G. (2017). Arctic microbiome and N-functions during the winter-spring transition. 1–68.
Taş, N., Prestat, E., Wang, S., Wu, Y., Ulrich, C., Kneafsey, T., Tringe, S. G., Torn, M. S., Hubbard, S. S., & Jansson, J. K. (2018). Landscape topography structures the soil microbiome in arctic polygonal tundra. Nature Communications, 9(1). https://doi.org/10.1038/s41467-018-03089-z
Tripathi, B. M., Kim, H. M., Jung, J. Y., Nam, S., Hyeon Tae Ju, Kim, M., & Lee, Y. K. (2019). Distinct taxonomic and functional profiles of the microbiome associated with different soil horizons of a moist tussock tundra in Alaska. Frontiers in Microbiology, 10(JUN). https://doi.org/10.3389/fmicb.2019.01442
Wang, J. L., Dragone, N. B., Avard, G., & Hynek, B. M. (2022). Microbial Survival in an Extreme Martian Analog Ecosystem: Poás Volcano, Costa Rica. Frontiers in Astronomy and Space Sciences, 9(January), 1–16. https://doi.org/10.3389/fspas.2022.817900
Weimer, B., & Rompato, G. (2009). Microbial biodiversity of Great Salt Lake, Utah. Natural Resources …, 15(1), 15–22. http://digitalcommons.usu.edu/cgi/viewcontent.cgi?...
Zhang, E., Thibaut, L. M., Terauds, A., Raven, M., Tanaka, M. M., Van Dorst, J., Wong, S. Y., Crane, S., & Ferrari, B. C. (2020). Lifting the veil on arid-to-hyperarid Antarctic soil microbiomes: A tale of two oases. Microbiome, 8(1), 1–12. https://doi.org/10.1186/s40168-020-00809-w