An Introduction to Isothermal Amplification
by Katharine Martin

by Katharine Martin
In molecular biology, when it comes to copying nucleic acid sequences, PCR gets all the glory. But there is another, less common term floating around: isothermal amplification, and maybe it’s got you wondering, “well, what is that exactly, and how is it different from PCR?” Isothermal amplification is the continuous, exponential amplification of nucleic acid sequences at a constant temperature using enzymes, usually strand displacing polymerases, rather than temperature changes.
That might seem pretty technical, so to break it down, isothermal amplification is a way of copying nucleic acids without temperature changing cycles. Instead, it uses specific DNA polymerases (usually), and specially designed primer sets to exponentially amplify a target sequence. And this technology has helped overcome certain limitations of PCR.
In this article, we’ll talk more about Isothermal amplification, the techniques used, how it differs from PCR, and more.
What is Isothermal Amplification
List of isothermal amplification techniques
The difference between isothermal amplification and PCR
Advantages of isothermal amplification
Why isothermal amplification is not as popularly used as PCR
First-to-market advantage of PCR
Speed is not always an advantage.
When is isothermal amplification appropriate to use?
Is Isothermal Amplification is Right for Your Lab Checklist
The advantages of isothermal amplification kits
One of the common ways in which isothermal amplification works is by using a DNA polymerase that has strand displacement activity. Polymerase strand displacement occurs when DNA polymerase begins to extend primers while displacing the downstream template.
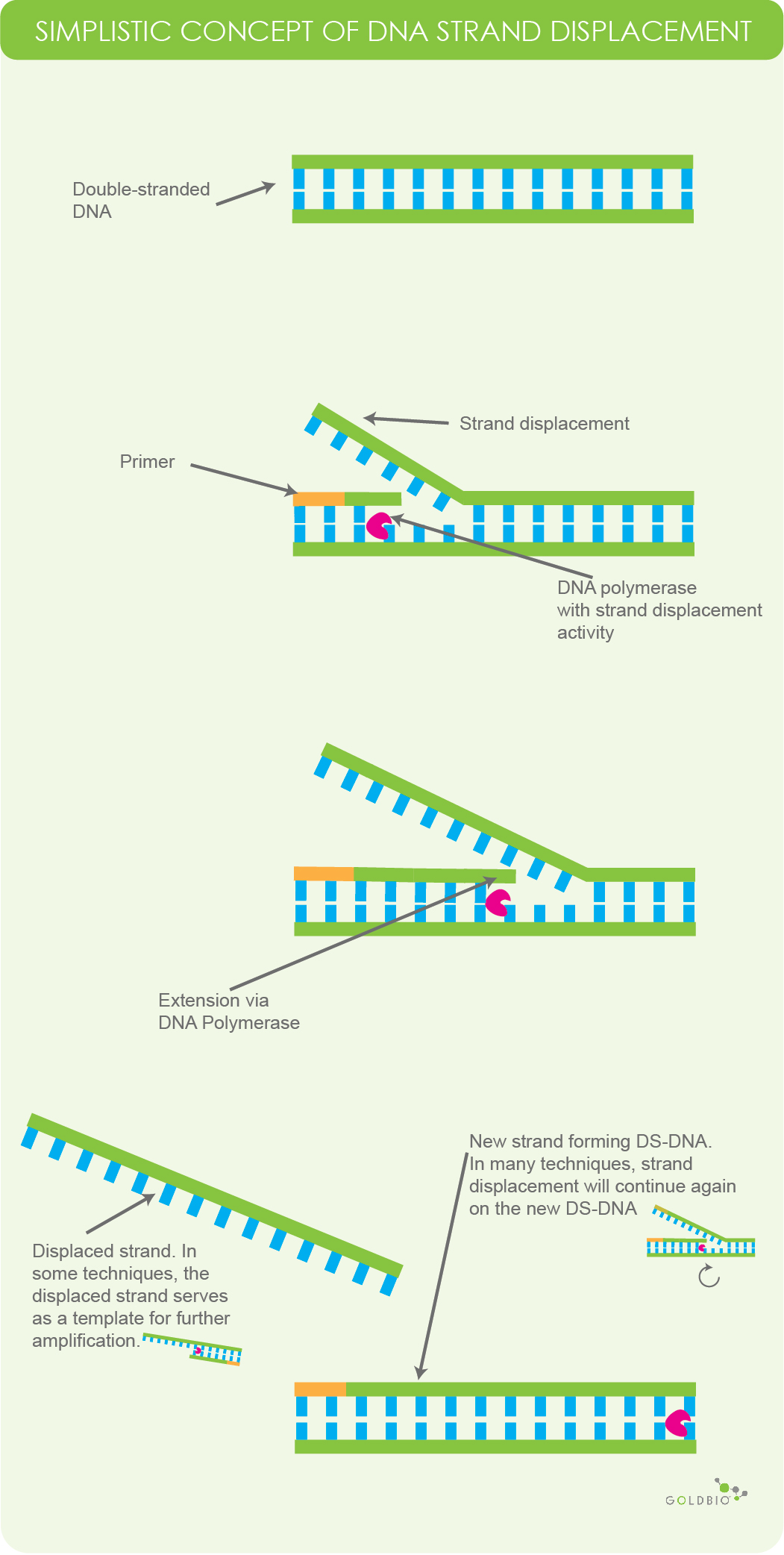
Figure 1. Simplistic overview of DNA polymerase with strand displacement activity. DNA polymerase will replicate a new strand of DNA while displacing one of the original strands. This process will continue repeatedly. And in some techniques, the displaced strand serves as a template for further amplification. Note, this figure is meant for conceptual understanding and does not represent all types of strand displacing behavior in isothermal amplification.
It is important for us to point out that when we talk about strand displacement, it’s in a very general context. Different variations employ different techniques and events, such as an initial nicking event, branching and the use of specialized primer sets.
There are also other isothermal amplification techniques that depend on other types of enzymes to unzip the target strand of DNA. For instance, helicase-mediated isothermal amplification uses T7 helicase to unwind DNA, allowing primer binding (Xu, Kim, Kays, Rice, & Kong, 2006).
Isothermal amplification is not a new technique. It has been around since the 1990s and has continued to evolve. As of now, there are several different approaches depending on amplification goals, some which will be discussed a little bit in this article. Among the techniques available are:
The key difference between isothermal amplification and PCR is isothermal amplification is carried out at a constant temperature using amplification machinery, whereas PCR requires temperature-changing cycles for amplification. What this also means is that isothermal amplification does not require a thermal cycler (though many machines can also perform isothermal amplification) for the reaction.
Another important difference between isothermal amplification and PCR is that DNA polymerase will extend primers on a single strand of DNA during PCR, while certain isothermal amplification techniques have special stand displacing DNA polymerases that extend primers on double-stranded DNA.
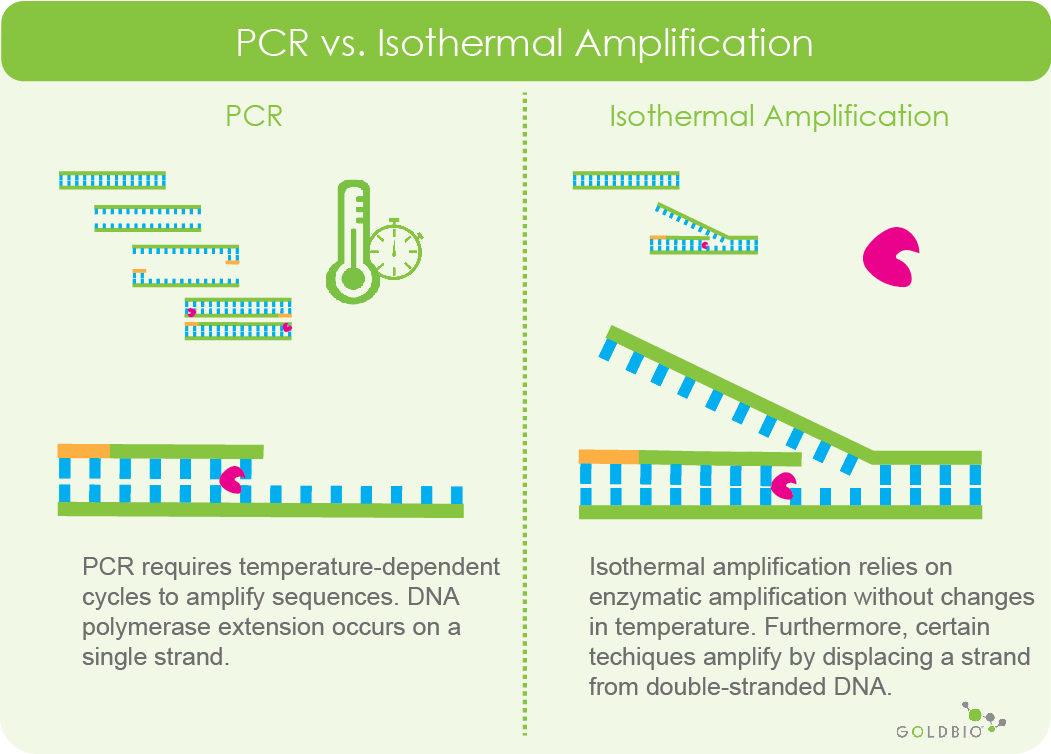
Figure 2. Comparing the basic differences between PCR and isothermal amplification. PCR requires repeated cycles of programmed temperature changes to exponentially amplify sequences. Isothermal amplification relies on enzymatic characteristics to carry out amplification. This example shows the very basic concept of isothermal amplification via strand displacement. In this comparison, DNA polymerase can extend a primer while displacing a strand of double-stranded DNA. However, for PCR, DNA polymerase extends a primer using a single DNA strand as a template.
One of the biggest advantages of isothermal amplification is that it doesn’t require thermal cycling, and therefore requires less power consumption, making it a compatible technique for hand-held equipment in the field.
Being able to carry out amplification on-site with handheld equipment opens the door for molecular biologists. They can perform point-of-care diagnostics or examine onsite environmental samples (Zanoli & Spoto, 2013).
The second advantage to isothermal amplification is its speed and sensitivity. Because isothermal provides continuous, exponential amplification independent of thermal cycling. Some methods can be performed in as little as 10 minutes (Zou, Mason, & Botella, 2020).
Depending on the technique used, primer design for isothermal amplification also enables greater target specificity (Zou, Mason, & Botella, 2020), which can be extremely useful, especially for whole genome amplification.
If isothermal amplification doesn’t require thermal cycling, can be carried out on-site, is fast and sensitive, why isn’t it as commonly used as PCR?
We’ve listed a few reasons isothermal amplification is not as popular as PCR:
PCR came just a little bit before isothermal amplification, and the science behind it was relatively easy to understand. At programed temperature intervals, the behavior of DNA, primers and polymerase could be manipulated into exponential amplification. Because it was first and easy to setup, it was quickly adopted by laboratories. From there, PCR continued to evolve. New techniques spun out fulfilling other research needs (Howard, 2019).
The LAMP method, as an example, requires six primers rather than the two needed for PCR. Primer design can be a little frustrating, especially when you’re new at it, reducing that frustration is going to be extremely attractive. If your lab is already set up for PCR, there would have to be a case for spending extra time and money developing and purchasing special primers (Howard, 2019).
Keep in mind, however, if you’re considering isothermal amplification for a regular diagnostic test, you may only have to worry about primer design once. Which means you would only encounter this frustration one time. Furthermore, kits can ease a lot of frustration since there is no need for primer design.
A general PCR master mix contains dNTPs, template, water, primers, Taq DNA polymerase, MgCl2 and reaction buffer. However, different types of isothermal amplification techniques require other enzymes, probes, proteins, etc. (Howard, 2019). Kits also succeed in overcoming this issue. Amplification kits will come with all the components needed for your reaction so you’re not stuck with extra reagents, or having to spend time shopping and price comparing.
With basic PCR technology having been available for a few decades now, there are fewer intellectual property barriers.
Older isothermal amplification methods have fewer barriers, but the new, innovative techniques that are more advantageous do (Howard, 2019).
Isothermal amplification happens quickly. Depending on the setup, results could be achieved in as little as 30 – 60 minutes. In some cases, results are produced in as little as 10 minutes. Yet speed is not always an advantage. For example, high-throughput batch production using PCR saves money and time with a simple process. And since production is scaled, these advantages far outweigh the speed obtained from isothermal amplification.
Thinking about some of the setbacks of isothermal amplification, you might wonder when is it right for your lab. When thinking about whether isothermal amplification is right for you, consider how important speed is to you, whether you need an extremely sensitive test, and if you need to perform the test on-site.
Deciding whether isothermal amplification would be a better approach for your lab involves a balance of the factors listed above. For instance, if you need a rapid, on-site test using the same diagnostic test every time, the advantages of isothermal amplification will become more critically important than the ease of PCR.
To help you better weight out these factors, we have provided a table for you to use when considering isothermal amplification.
Is Isothermal Amplification is Right for Your Lab Checklist |
|
|
Do you need results in under an hour? |
Yes / No |
|
Do you need to perform on-site testing? |
Yes / No |
|
Do you have a smaller lab (not doing large-batch amplification)? |
Yes / No |
|
Are you doing a regular diagnostic test that would use the same primers? |
Yes / No |
|
Are you working with small sample amounts needing high sensitivity? |
Yes / No |
|
Do you have equipment that will carry out isothermal amplification? (Many thermal cyclers can carry out the reaction.) |
Yes / No |
|
Do you have the budget for the components listed your intended protocol? |
Yes / No |
The more YESes checked off on this list (with the exception of the last two questions, whose NOs may in fact be deal breakers), the more likely isothermal amplification would be appropriate for your lab.
If you discover isothermal amplification is critical for your laboratory, you might not want to deal with the frustration of selecting which type of amplification method to use, designing primers, shopping for components and optimizing the test.
If you’re running a standard diagnostic, this is where kits provide an excellent benefit. With a kit, you can be assured your setup is optimized and you have all the components you need. And if you’re nervous about ease of use, kits typically make reaction setup extremely user-friendly.

DNA gel electrophoresis is one of the most widely used analytical techniques in molecular biology, providing a simple and reliable way to separate DNA fragments...

Nickel affinity chromatography is a technique used to purify proteins that contain a polyhistidine tag (his-tag). Nickel chromatography is a popular choice for affinity purifications...

The best agarose for DNA gels depends on your application. Agarose LE is ideal for routine electrophoresis. Low melt agarose is best for DNA recovery,...

Making IPTG stock solution involves weighing out IPTG powder, pouring it into a conical tube or cylinder, and adding deionized water to the desired volume....
