Human Infections and the Microbiome
by Adriana Gallego, Ph.D.

by Adriana Gallego, Ph.D.
When people feel sick, they usually rest to recover. If it’s very severe, a person will schedule see their doctor, and then the doctor will identify the infection and come up with a treatment plan.
In the absence of knowledge about the specific pathogen causing the infection, broad-spectrum antimicrobial drugs, which kill many different microbe species at once, are prescribed.
Although this approach saves many lives by fighting infections, the current knowledge base on the microbiome has led to a paradigm change without precedents.
We moved from a hypothesis established 150 years ago with Koch’s postulates where microbes were primarily recognized as the cause of significant illnesses towards one where microbes are good for us.
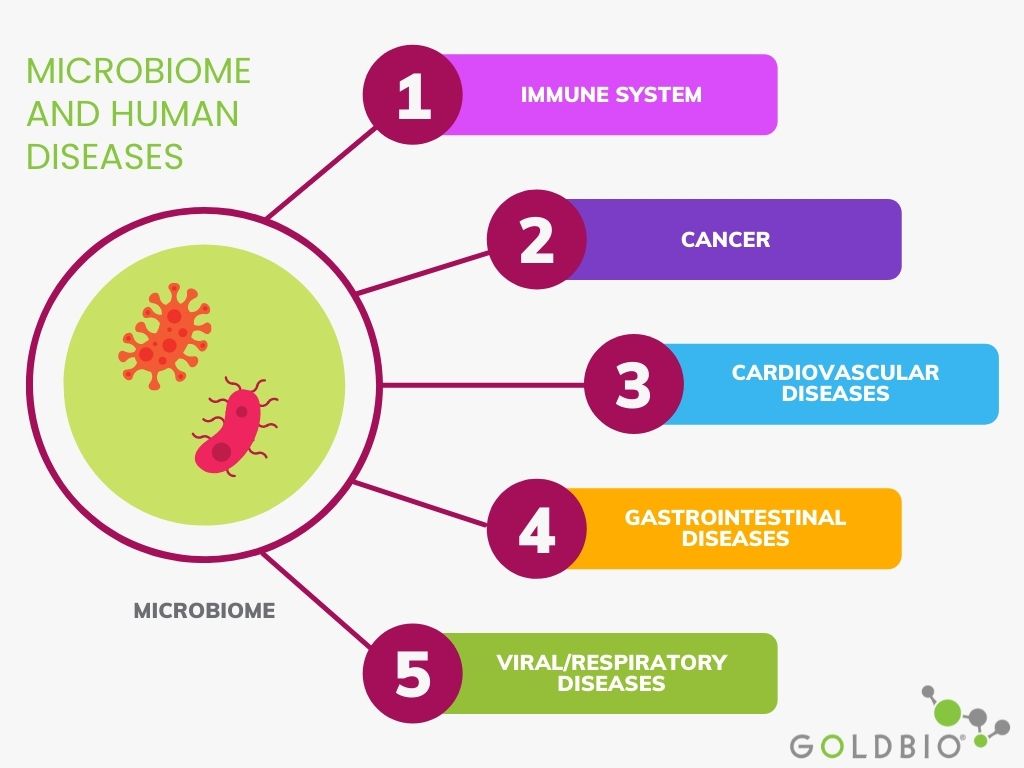
In this article, I will explain the relationship between the microbiome and different types of human diseases and some microbiome-based therapies to fight against various infections.
Graphical abstract
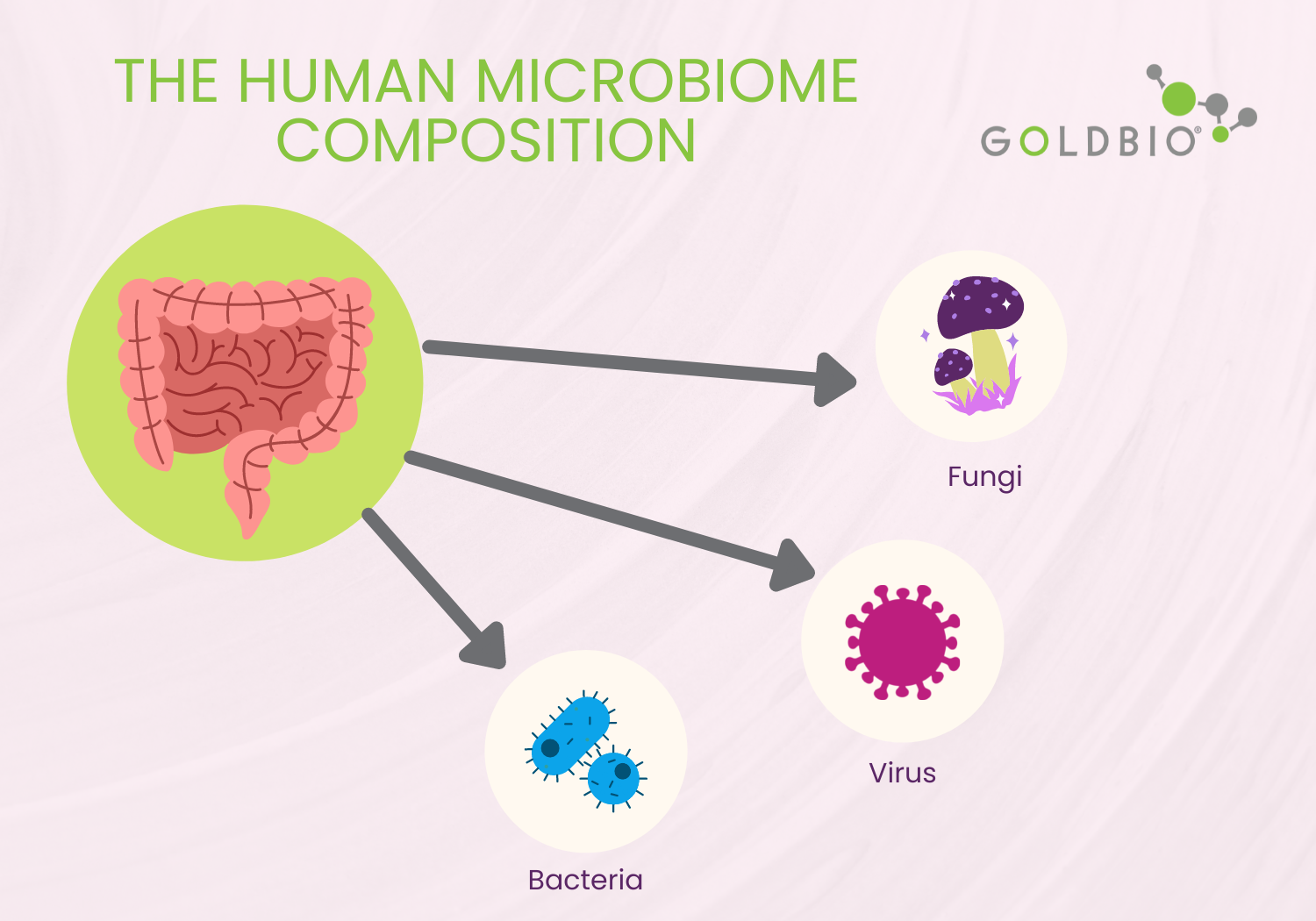
The human microbiome consists of bacteria, viruses, and fungi. They are ideally living symbiotically within their host. However, bacteria (bacteriome) remain the most studied group within the human microbiome.
When we talk about diseases, we must also discuss the concept of dysbiosis.
Dysbiosis is defined as any change to the composition of microbial communities relative to those found in healthy individuals.
For instance, dysbiosis occurs when there is an increase in potential pathogenic class such as Gammaproteobacteria.
Dysbiosis can also be a result of a decrease in beneficial microbes like Bifidobacterium species, or when there is a change in the richness or depletion of a species in a given microbiome.
Although dysbiosis remains a cause or consequence of changes in human physiology and disease, today dysbiosis is a benchmark to compare healthy and sick individuals.
There are two types of human immune systems. One is called the innate immune system, and the other is the adaptative immune system.
The innate immune system is the immediate defense mechanisms responding to pathogen exposure.
The innate immune system comprises the following:
Innate leucocytes are a defense mechanism and they are pathogen-specific. Some innate leucocytes include antibody-producing B-cells and T-cells. These cells are able to act when the pathogen attacks a second time. This ability is also considered to be a pathogen-specific memory (Harper et al. 2021).
In other words, the innate immune system is the default defense mechanism, and it is immediately activated once our body detects the pathogen.
The second type of immune system is the adaptative immune system.
The adaptive immune system is activated during the infection to strengthen the responses given by the innate immune system. Consider it the second-line of defense.
One impressive advantage of the adaptative immune system is that it provides the body a memory of the pathogen.
Therefore, if the pathogen attacks a second time, the body will have the tools (antibodies) to stop the pathogenic attack.
A microbiome exerts its effects and modulates the responses to certain diseases based on the interplay, particularly innate immunity.
These responses can be modulated by activating specific genes or generating signaling pathways initiated by bacterial metabolic products.

There are several studies relating microbiomes and cancer. For instance, when dysbiosis in the gut occurs due to antimicrobial drugs or persistent infections, that could lead to an increased risk of colorectal cancer. Furthermore, the presence of H. pylori has been associated with the risk of gastric cancer in humans. In addition, Fusobacterium and Clostridium are also species overrepresented in individuals with gastric cancer.
Regarding breast cancer, Bacillus and Staphylococcus are more likely present in sick women than healthy individuals. Also, prostate cancer is related to a higher population of Bacteroides massiliensis. Still, researchers are still trying to determine if the presence of these microbes is a consequence or cause in the prevalence of cancer. What is clear is that these microbes are likely correlated with breast cancer.
Altogether, the alteration in the human microbiota in different body sites contributes to complex interactions between cancer and the human microbiota.

Some microorganisms in the gut microbiota produce trimethylamine N-oxide (TMAO) metabolites related to heart diseases.
These TMAO metabolites affect lipid transportation in the body and induce the release of precursors promoting foam cell formation and hardening of the arteries in animal models. It suggests that the accumulation of these metabolites may induce artery hardening, however more long-term research is needed to clarify this hypothesis.
Furthermore, a higher ratio of Firmicutes to Bacteriodetes species in the stool of hypertensive mice has been observed. And, a disrupted intestinal flora was associated with individuals having a higher risk of cardiovascular disease than healthy individuals (Ogunrinola et al. 2020).

The gastrointestinal (GI) microbiome is maybe one of the most studied microbiomes in humans. The GI microbiome plays a significant role in the host immune system and regulates defense mechanisms.
Dysbiosis in the gut is highly related with diseases of the GI system such as inflammatory bowel disease (IBD) and many other diseases such as cancer, cardiovascular, and respiratory infections.
Furthermore, systemic diseases are caused by the movement of bacteria from the intestinal mucosa to other extraintestinal sites. It allows the transfer of these organisms and toxins from the gut to other parts of the body, which induces inflammatory responses that trigger several diseases, including appendicitis.

Although several viral diseases have caused the death of millions of people throughout the history of humanity, with the COVID-19 outbreak, attention to the human microbiome and its relationship with viral diseases increased significantly.
Many viral diseases cause acute respiratory distress syndrome (ARDS), such as influenza, MERS-CoV, and SARS-CoV infections.
For patients with ARDS and the associated dysbiosis, there is a positive correlation with alveolar intensity (the sponge part of the lung) and systemic inflammation.

Interestingly, the main organ affected by many viral diseases is the lung. Although it was thought that lungs were germ-free organs, with microbiome studies, research found that microbes live in the lung naturally (Cyprian et al., 2021). These microbes are shaped by a balanced gut microbiome that affects the mucosa of the lungs.
For instance, Ji et al. (2021) gave a set of bacteria (Escherichia coli, Streptococcus thermophilus, Bifidobacterium spp., and Lactobacillus spp.) to baby mice. It suppressed infection with a respiratory syncytial virus (RSV) and protected the host against the lung disease caused by the virus. This defense response was associated with the abundance of Corynebacterium and Lactobacillus in the lungs.
One interesting thing derived from studies on farming environments is that a high microbial diversity is connected to lower reports of respiratory diseases.
One mechanism behind this phenomenon has been linked to the activation of the innate immune system in the epithelial cells of the respiratory tract (Ogunrinola et al. 2020).
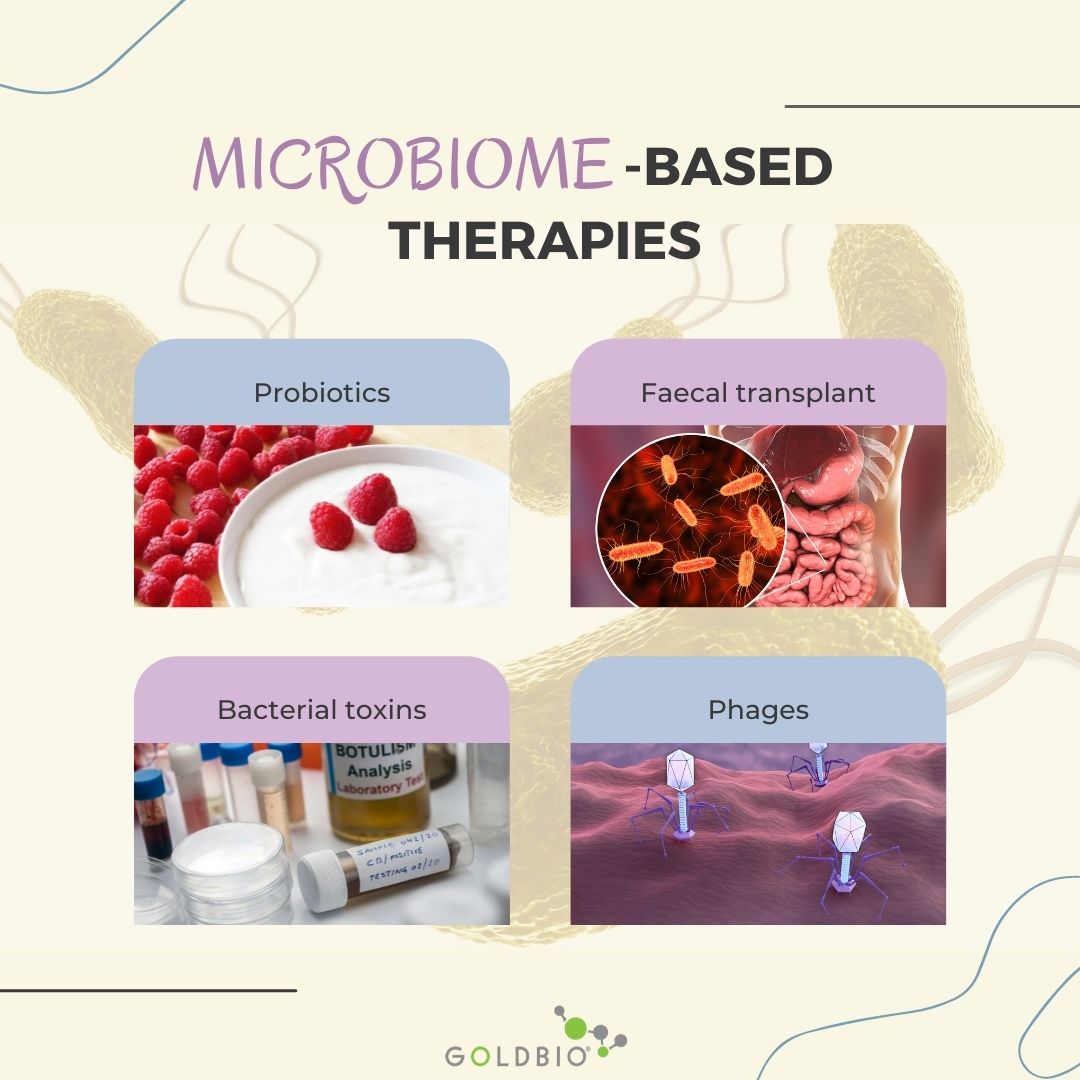
Several anti-infection strategies based on the microbiome knowledge range from using an entire intact microbiome or a microbial consortium to selecting single biological entities (bacteria, fungi, bacteriophages, or bacterial metabolites).
Four strategies were reported to be based on microbiome knowledge. Probiotics, fecal transplant, and bacterial toxins and phages use.
Probiotics are live microorganisms that confer a health benefit on the host (Hill, 2021).
One significant study showed that 4,500 children consumed Lactiplantibacillus plantarum together with fructooligosaccharides. The children were at least 2,000 g at birth, at least 35 weeks of gestation, and with no signs of sepsis or other morbidity, and they were monitored for 60 days. The study showed a significant reduction in respiratory tract infections (Panigrahi et al., 2017).
A fecal transplant involves collecting the entire microbiome from one healthy individual using his or her stool and transplanting it with minimal processing to the colon of a patient with an infection. This fecal transplant has been successful in the treatment of Clostridium difficile infections.
In other cases, spores purified from a fecal sample have been transferred to sick patients.
Although this intervention had less success in phase 2 clinical trials, it holds the potential to be part of a range of microbiome-based intervention strategies (O´Hara et al., 2006).
Bacteriocins are a type of bacterial toxin. Here you can find broad and narrow spectrum bacteriocins.
Broad-spectrum toxins have similar effect as antibiotics. A high amount of antibiotics in the human microbiome can kill different bacterial species at once creating dysbiosis. Therefore, when possible, it is better to use narrow-spectrum bacteriocins to avoid dysbiosis events.
In this sense, thuricin CD, is a very narrow-spectrum bacteriocin targeting C. difficile. Because bacteriocins are encoded genes, they can be engineered to optimize their physicochemical features and reduce their spectrum. Furthermore, bacteriocins can also be used in combination with other antimicrobials to have a greater effect.
Phages are bacterial viruses that often have a very narrow-spectrum of inhibition. Yes, even phages fall within a microbiome.
Unfortunately, when it comes to microbiome-based therapies, this approach has disadvantages such as the ease with which bacteria can develop resistance to phage attacks. But there are also advantages, such as their widespread distribution in nature, favoring subsequent isolation in many instances, and their ability to multiply at the site of infection.
Furthermore, lysins are enzymes produced by bacteriophages. They are responsible for lysing the cell wall of the target bacterium avoiding the multiplication of phage particles.
Some efforts have been reported to use lysins to target specific pathogens, particularly on the skin and respiratory tract.
One of the most interesting aspects of these microbiome-based inhibitors is their potential narrow-spectrum of inhibition, which would selectively deplete the pathogen while preserving the complexity and richness of the surrounding microbiome. Moreover, the microbiome knowledge will boost the development of pathogen-specific strategies that improve humans' well-being and immune system.
Microbiome, human diseases, immune system, microbiome-based therapies.
Amato, K. R., Azad, M., Blaser, M. J., Thomas, C. G., Chu, H., Dominguez-bello, M. G., Dusko, S., Elinav, E., Geva-zatorsky, N., Gros, P., Keck, F., Korem, T., Mcfall-ngai, M. J., Melby, M. K., Nichter, M., Pettersson, S., Rees, T., Tropini, C., Zhao, L., … Bosch, T. C. G. (2021). The hygiene hypothesis , the COVID pandemic , and consequences for the human microbiome. 118(11). https://doi.org/10.1073/pnas.2102333118
Cyprian, F., Umar, M., Abdelhafez, I., Salman, S., Attique, Z., Kamareddine, L., & Al-asmakh, M. (2021). International Journal of Infectious Diseases SARS-CoV-2 and immune-microbiome interactions : Lessons from respiratory viral infections. International Journal of Infectious Diseases, 105, 540–550. https://doi.org/10.1016/j.ijid.2021.02.071
Gonzalez, R., & Elena, S. (2021). The Interplay between the Host Microbiome and Pathogenic. MBio, 12(6), 1–13.
Harper, A., Vijayakumar, V., Ouwehand, A. C., Haar, J., Obis, D., Espadaler, J., Binda, S., Desiraju, S., & Day, R. (2021). Viral Infections , the Microbiome , and Probiotics. 10(February), 1–21. https://doi.org/10.3389/fcimb.2020.596166
Hill, C. (2021). Microbiome and Infection : A Case for “ Selective Depletion .” 77(suppl 3), 4–9. https://doi.org/10.1159/000516399
Ji, J. jian, Sun, Q. mei, Nie, D. yun, Wang, Q., Zhang, H., Qin, F. fen, Wang, Q. sheng, Lu, S. feng, Pang, G. ming, & Lu, Z. gang. (2021). Probiotics protect against RSV infection by modulating the microbiota-alveolar-macrophage axis. Acta Pharmacologica Sinica, 42(10), 1630–1641. https://doi.org/10.1038/s41401-020-00573-5
O’Hara, A. M., & Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Reports, 7(7), 688–693. https://doi.org/10.1038/sj.embor.7400731
Ogunrinola, G. A., Oyewale, J. O., Oshamika, O. O., & Olasehinde, G. I. (2020). The Human Microbiome and Its Impacts on Health. International Journal of Microbiology, 2020. https://doi.org/10.1155/2020/8045646
Panigrahi, P., Parida, S., Nanda, N. C., Satpathy, R., Pradhan, L., Chandel, Di. S., Baccaglini, L., Mohapatra, A., Mohapatra, S. S., Misra, P. R., Chaudhry, R., Chen, H. H., Johnson, J. A., Morris, J. G., Paneth, N., & Gewolb, I. H. (2017). A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature, 548(7668), 407–412. https://doi.org/10.1038/nature23480
Ranallo, R. T., Mcdonald, L. C., Halpin, A. L., Hiltke, T., & Young, V. B. (2021). The State of Microbiome Science at the Intersection of Infectious Diseases and Antimicrobial Resistance. 223(Suppl 3), 187–193. https://doi.org/10.1093/infdis/jiab020
Schwartz, D. J., Langdon, A. E., & Dantas, G. (2021). Understanding the impact of antibiotic perturbation on the human microbiome. 2020, 1–12.

A His-tag is a stretch of 6-10 histidine amino acids in a row that is used for affinity purification, protein detection, and biochemical assays. His-tags...

Competent cells such as DH5a, DH10B, and BL21 will maintain their transformation efficiency for at least a year with proper storage. It is important to...

Ni2+ ions give nickel agarose beads their characteristic blue color. This blue color can fade or disappear completely when loading his-tagged proteins onto the column....

Nickel agarose beads change from blue to a brown or black color when the nickel ions have been reduced from a Ni2+ to a Ni1+...
