The 5 Golden Tips for Library Preparation
by Adriana Gallego, Ph.D.

by Adriana Gallego, Ph.D.
Being successful at preparing a library is always tricky because there are many factors involved. They include the input material, the chosen library kit, the sequencing goal, and many others.
With all of those factors in mind, I have put together a detailed list of the top 5 tips for library preparation. These tips are logical and sequential to help you at each step of the process.
Article Table of Contents
The routes for library prep: Which to choose?
Tip 2: Pay attention to the quality of your input material
Tip 3: Selecting a kit for library prep
Methods in Next-Generation Sequencing
Systems in Next-Generation Sequencing
Tip 4: Follow good laboratory practices
Tip 5: Check, check, and check
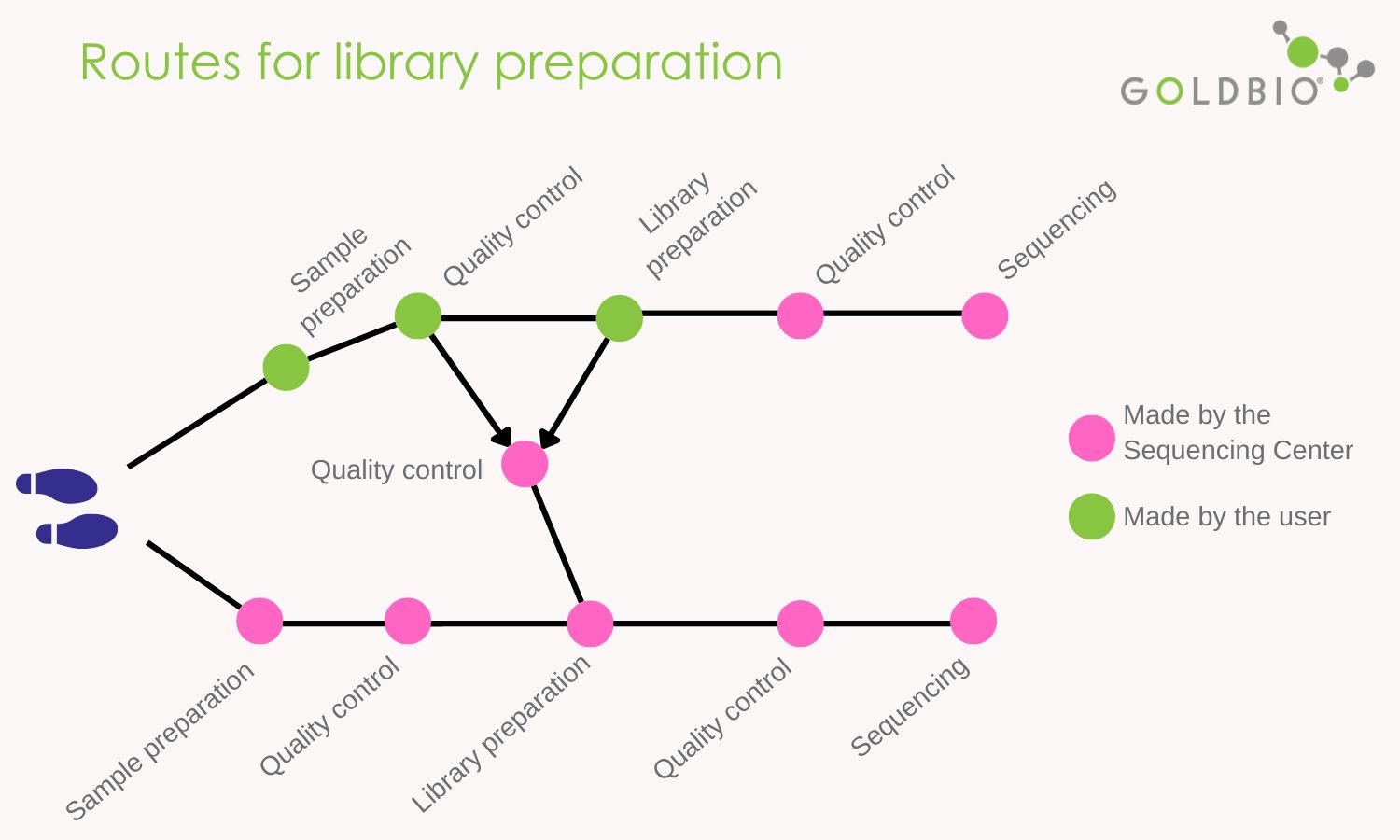
You can make your library prep or pay a sequencing center to do it. Each approach has pros and cons, so let me give you some indications about choosing either one of the routes.
For instance, when the scientist prepares her libraries, she benefits from learning by doing and can identify critical checkpoints during library prep. Also, and no less important, preparing her own libraries reduce costs significantly.
The downside is that the initial learning curve is time-consuming. Also, if the process fails, researchers can incur extra expenses for reagents and consumables, and samples can be lost.
When you hire a sequencing center to prepare samples for you, you benefit from the automatized options these centers have. They also use less time for processing, and high-quality libraries are obtained.
Furthermore, these centers perform quality controls at different points of the library prep, guaranteeing a clean and systematic execution of the process. Technicians at the centers may guide you in your NGS needs.
The disadvantage, in this case, is the cost which can be significantly higher compared to consumer-made preparations. This cost can be even higher if you do not provide high-quality DNA.
With that in mind, let’s proceed with the top five tips for library preparation.
Whether you are new at this or a seasoned researcher, NGS is not a big deal when you have a clear sequencing scope.
NGS applications such as whole genome sequencing (WGS), RNA sequencing (RNA-Seq), methylation-sequencing, exome-sequencing, metagenomics, and so on have specialized library kit options.
For instance, RNA-Seq library kits look to enrich your mRNA or deplete your rRNA. And exome-Seq kits look for exons that can be captured with specific adapters.
So, with so many kit options it is important you establish the scope of your NGS project and ask yourself about the application, project size and available budget.
For example, which organism are you working with? How many samples are you planning to prepare and sequence? How much are you planning to spend on the NGS project? How much time do you have to prepare the libraries? Are you a beginner or advanced in library prep? These questions need to be answered before proceeding with library prep.
Check our Library prep questionnaire to help you be prepared for running an NGS project.
Setting your scope will help you choose the proper kit and what you may expect in terms of time and costs.
Whether you decide to prepare the samples on your own or pay for the preparation, it is critical to start with high-quality genomic DNA. A low-quality input sample (e.g., contamination) interferes with the performance of library preparation and can skew the results.
If you don’t check the quality of your input DNA carefully, you can be in a situation where samples are returned from the sequencing center because they failed the quality controls, which wastes time and money.
Available systems to verify the quality of your input material are:
Nanodrop: a system that measures the amount of DNA/RNA present in your sample using spectrophotometry. It is based on the ratios 260/280 and 260/230. The maximal absorption for DNA is at 260nm.
So, the ratios are used as indicators of contamination. If 260/280 is lower than 1,8, the DNA sample is considered to have high purity. If the absorbance at 260/230 is lower than 1,8, it means there is possible contamination with organic compounds like polyphenols.
QubitTM: is a fluorescence-based system where fluorescent dyes are bound to the DNA. Here, the amount of DNA is proportional to the fluorescence intensity. QubitTM is used to determine the concentration of DNA.
Bioanalyzer: a system based on electrophoresis that measures the concentration and quality of nucleic acids and proteins. It is very accurate, making it a great option for library preparation when possible.

Selecting the proper library prep kit may be challenging because there are about 114 kits available for NGS applications.
So, how do you choose? My recommendation is after the selection of application and species, you can move into the methods and the systems.
Methods in NGS are indeed the procedures to perform a given application. There are two types of methods such as those based on sequencing and those based on arrays.
Systems instead refers to the type of sequencers. Let’s take a closer look at each.
Methods based on sequencing are procedures that do not depend on previous knowledge of the organism - that is, you don’t need a reference genome, and they are ideal for discovery.
Instead, methods based on arrays require prior knowledge of the organism and designed probes. It makes arrays ideal for profiling approaches.
For methods sequencing-based you can find:
For methods array-based you can find:
Methods based on sequencing are commonly used in applications such as metagenomics, whole genome sequencing or RNA sequencing (RNA-Seq).
Instead, methods based on arrays are used in applications like Methylation-Seq and Exome-Seq.
Systems are essentially the type of sequencers. They include the series of HiSeq® (1000, 1500, 2000, 2500, 3000, 4000, X Five), MiSeq® (MiSeq and Dx), and NextSeq® (500, 550, 550Dx, 1000, 2000), and other systems like NovaSeq®, MiniSeq®, and iSeq®.
Differences between these sequencers include the throughput and the amount of data they can provide. But no worries! You can easily ask your sequencing center what sequencers they have available and request a recommendation based on the amount of data you want to get.
Altogether, to help you select the right library prep kit, you can use a handy online tool called the Library Prep and Array Kit Selector for free. Play around!

Although following a kit may appear very simple, sometimes, researchers forget the basic steps in molecular biology. Keep in mind that preparing a library requires extra care because several downstream steps (e.g. sequencing, data interpretation and qPCR validation) may also fail if you don’t prepare a good library.
Basic good lab practices to remember:
Quality control should be done before DNA use and after library preparation. At sequencing centers, additional quality controls are performed on the library when you submit your samples.
Quality control is a step used to check the quantity and quality of the libraries.
The techniques used in quality control of a library prep are:
The qPCR method is precise and usually more affordable for most molecular laboratories.
Fluorometrics, such as the Qubit system, is easy to use, but not all labs have one.
Finally, microfluidics methods like Bioanalyzer are accurate but usually require specialized technicians. Also, you can only analyze a low amount of samples at the same time. Furthermore, Bioanalyzer can be more expensive compared to the other two systems.
Library prep, tips, NGS goals, library prep selection, quality control.
Hadfield, J. (2014). Choosing an NGS library prep kit provider. Enseqlopedia.com. Retrieved from http://enseqlopedia.com/2014/04/choosing-an-ngs-library-prep-kit-provider/.
Head, S. R., Komori, H. K., LaMere, S. A., Whisenant, T., Van Nieuwerburgh, F., Salomon, D. R., & Ordoukhanian, P. (2014). Library construction for next-generation sequencing: Overviews and challenges. BioTechniques, 56(2), 61-77. https://doi.org/10.2144/000114133
Hess, J. F., Kohl, T. A., Kotrová, M., Rönsch, K., Paprotka, T., Mohr, V., Hutzenlaub, T., Brüggemann, M., Zengerle, R., Niemann, S., & Paust, N. (2020). Library preparation for next generation sequencing: A review of automation strategies. Biotechnology Advances, 41, 107537. https://doi.org/10.1016/j.biotechadv.2020.107537
Song, Y., Milon, B., Ott, S., Zhao, X., Sadzewicz, L., Shetty, A., Boger, E. T., Tallon, L. J., Morell, R. J., Mahurkar, A., & Hertzano, R. (2018). A comparative analysis of library prep approaches for sequencing low input translatome samples. BMC Genomics, 19(1), 696. https://doi.org/10.1186/s12864-018-5066-2
Tvedte, E. S., Michalski, J., Cheng, S., Patkus, R. S., Tallon, L. J., Sadzewicz, L., Bruno, V. M., Silva, J. C., Rasko, D. A., & Dunning Hotopp, J. C. (2021). Evaluation of a high-throughput, cost-effective Illumina library preparation kit. Scientific Reports, 11(1), 15925. https://doi.org/10.1038/s41598-021-94911-0
Vecera, M., Sana, J., Oppelt, J., Tichy, B., Alena, K., Lipina, R., Smrcka, M., Jancalek, R., Hermanova, M., Kren, L., & Slaby, O. (2019). Testing of library preparation methods for transcriptome sequencing of real life glioblastoma and brain tissue specimens: A comparative study with special focus on long non-coding RNAs. PLOS ONE, 14(2), e0211978. https://doi.org/10.1371/journal.pone.0211978
Zhao, S., Zhang, C., Mu, J., Zhang, H., Yao, W., Ding, X., Ding, J., & Chang, Y. (2020). All-in-one sequencing: An improved library preparation method for cost-effective and high-throughput next-generation sequencing. Plant Methods, 16(1), 74. https://doi.org/10.1186/s13007-020-00615-3

A His-tag is a stretch of 6-10 histidine amino acids in a row that is used for affinity purification, protein detection, and biochemical assays. His-tags...

Competent cells such as DH5a, DH10B, and BL21 will maintain their transformation efficiency for at least a year with proper storage. It is important to...

Ni2+ ions give nickel agarose beads their characteristic blue color. This blue color can fade or disappear completely when loading his-tagged proteins onto the column....

Nickel agarose beads change from blue to a brown or black color when the nickel ions have been reduced from a Ni2+ to a Ni1+...
